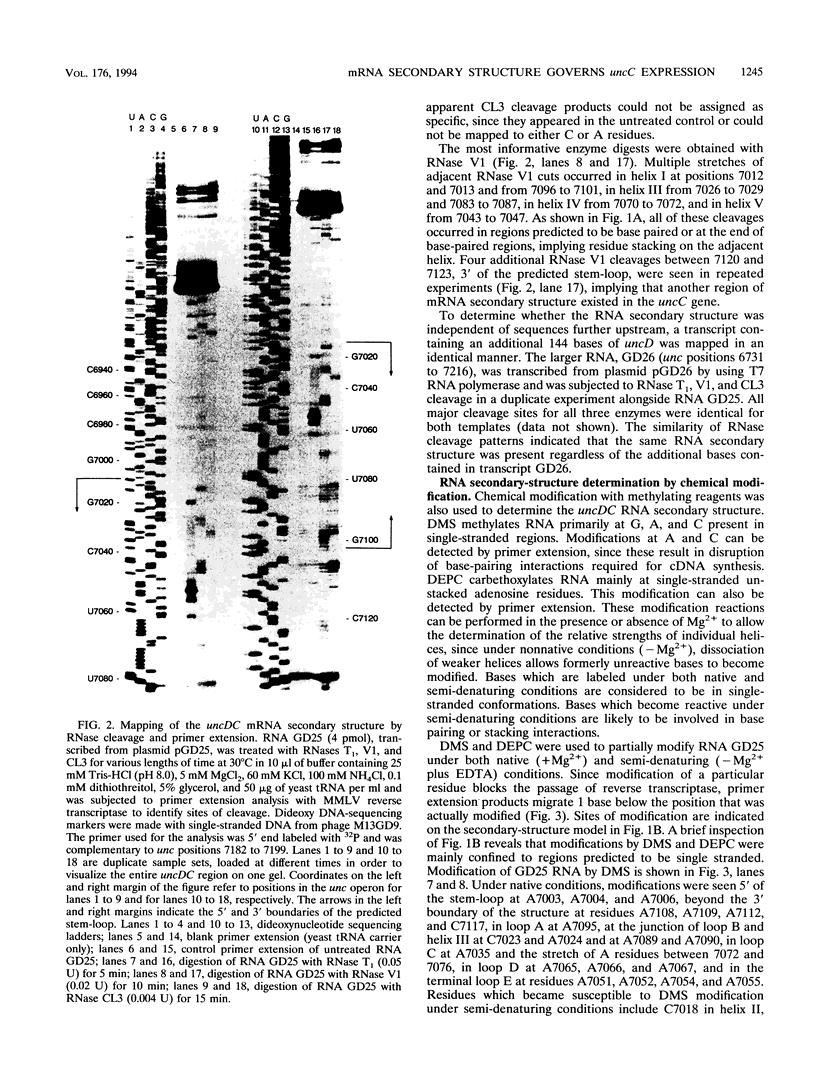
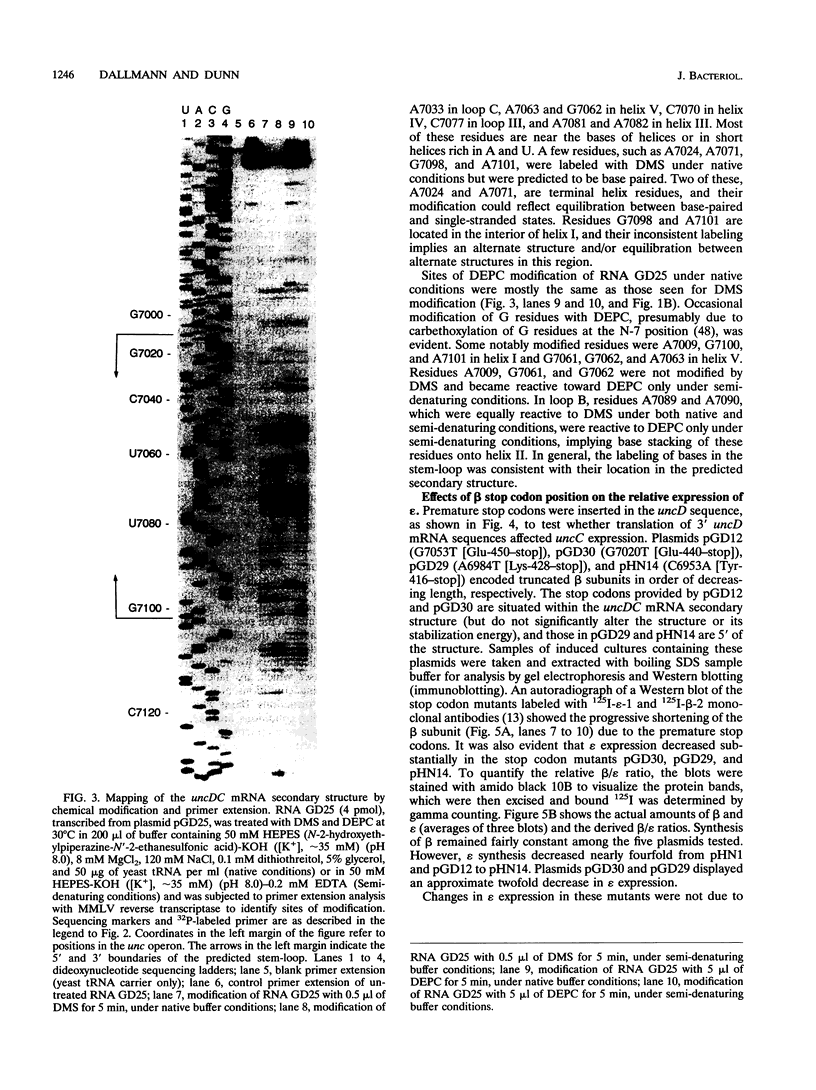
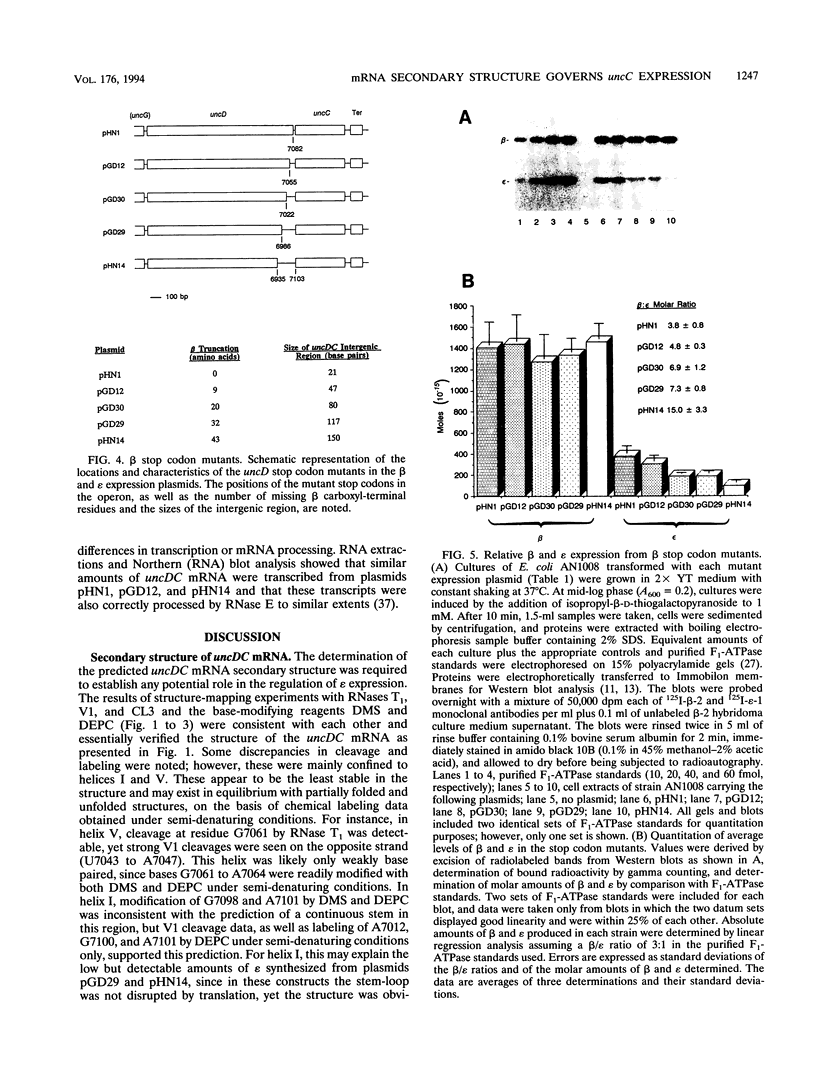
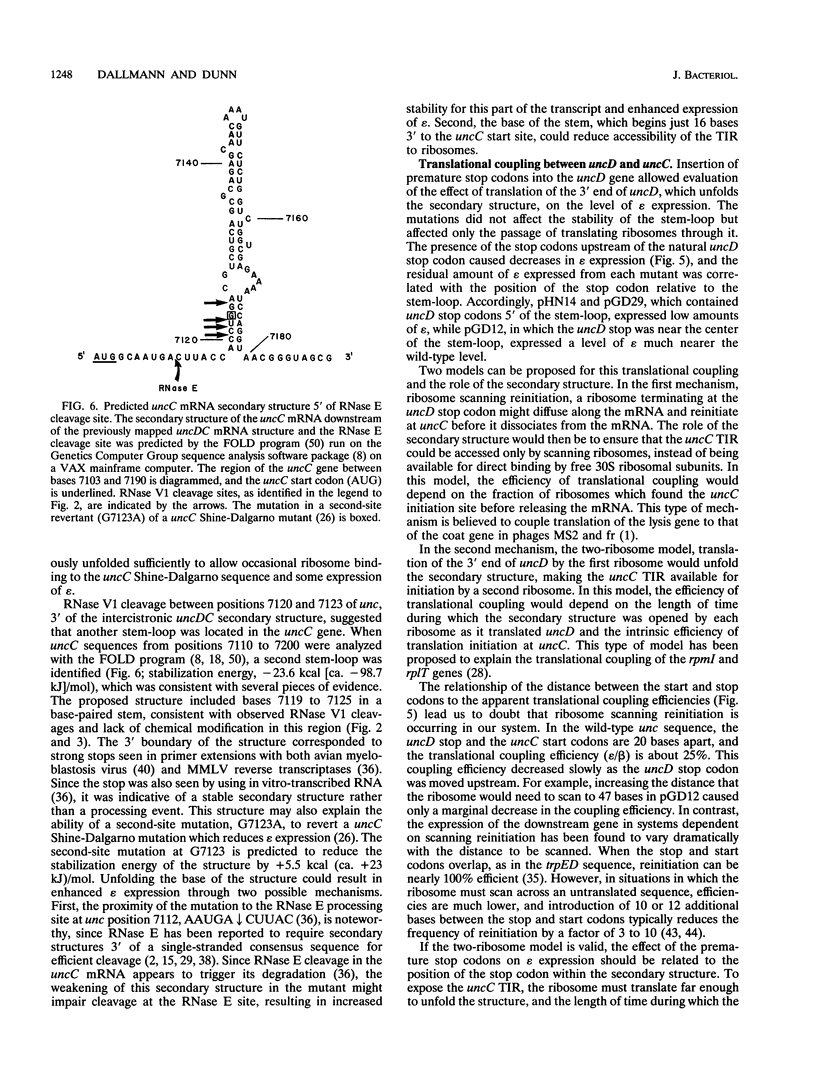
Abstract
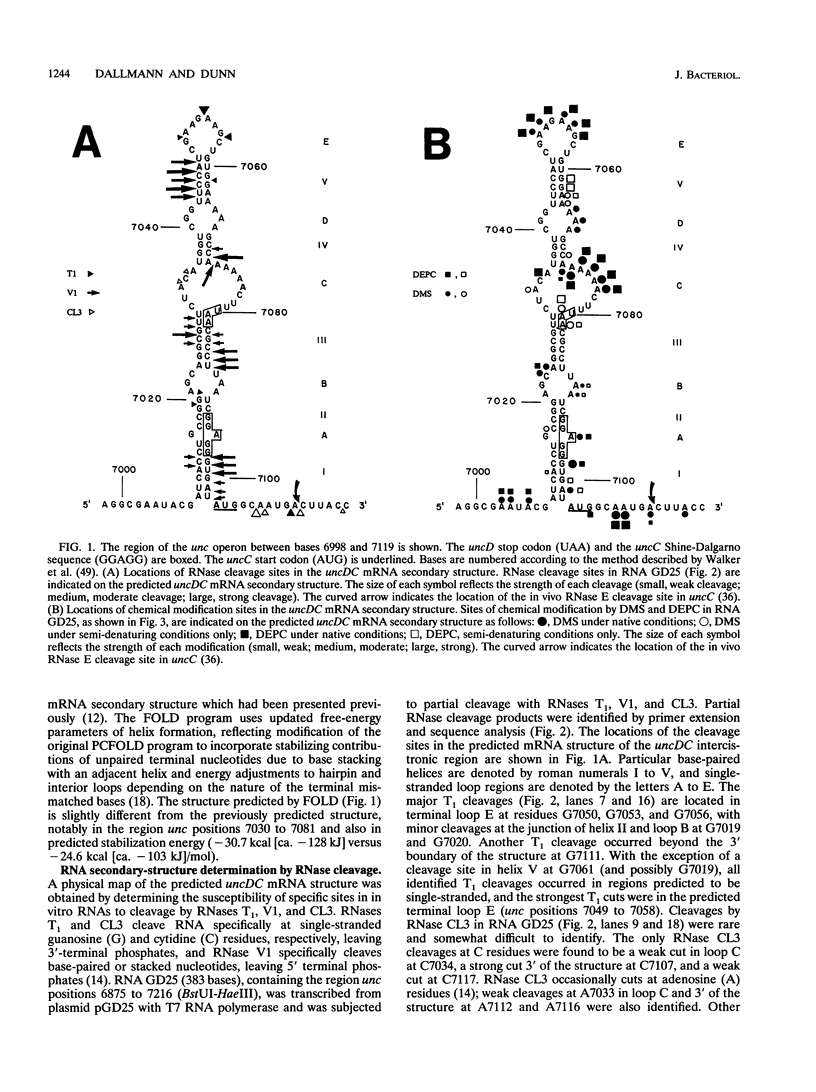
Escherichia coli expresses the beta and epsilon subunits of F1F0-ATP synthase at relative levels consistent with the 3:1 (beta/epsilon) stoichiometry in the holoenzyme. The mechanism of translational control of expression of the uncC gene (epsilon subunit) relative to the immediately 5' uncD gene (beta subunit) was examined. Previous expression studies and a computer analysis suggested the presence of an RNA secondary structure including the 3' end of uncD, the uncDC intergenic region, and the uncC Shine-Dalgarno sequence (S. D. Dunn and H. G. Dallmann, J. Bacteriol. 172:2782-2784, 1990). Analysis of in vitro-transcribed RNA by cleavage with RNases T1, V1, and CL3 and by chemical modification with dimethyl sulfate and diethyl pyrocarbonate confirmed a predicted structure. Introduction of premature uncD stop codons inserted 5' of the secondary structure strongly reduced epsilon expression, whereas stop codons inserted at positions within the secondary structure showed smaller effects, indicating that translational control of epsilon synthesis involves partial coupling to beta synthesis. Possible mechanisms by which the RNA secondary structure and the unfolding of this structure by translation of uncD may govern the level of uncC expression are discussed.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adhin M. R., van Duin J. Scanning model for translational reinitiation in eubacteria. J Mol Biol. 1990 Jun 20;213(4):811–818. doi: 10.1016/S0022-2836(05)80265-7. [DOI] [PubMed] [Google Scholar]
- Bouvet P., Belasco J. G. Control of RNase E-mediated RNA degradation by 5'-terminal base pairing in E. coli. Nature. 1992 Dec 3;360(6403):488–491. doi: 10.1038/360488a0. [DOI] [PubMed] [Google Scholar]
- Brusilow W. S., Klionsky D. J., Simoni R. D. Differential polypeptide synthesis of the proton-translocating ATPase of Escherichia coli. J Bacteriol. 1982 Sep;151(3):1363–1371. doi: 10.1128/jb.151.3.1363-1371.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter P. Improved oligonucleotide-directed mutagenesis using M13 vectors. Methods Enzymol. 1987;154:382–403. doi: 10.1016/0076-6879(87)54086-1. [DOI] [PubMed] [Google Scholar]
- Dallmann H. G., Flynn T. G., Dunn S. D. Determination of the 1-ethyl-3-[(3-dimethylamino)propyl]-carbodiimide- induced cross-link between the beta and epsilon subunits of Escherichia coli F1-ATPase. J Biol Chem. 1992 Sep 15;267(26):18953–18960. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Downie J. A., Langman L., Cox G. B., Yanofsky C., Gibson F. Subunits of the adenosine triphosphatase complex translated in vitro from the Escherichia coli unc operon. J Bacteriol. 1980 Jul;143(1):8–17. doi: 10.1128/jb.143.1.8-17.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dreyfus M. What constitutes the signal for the initiation of protein synthesis on Escherichia coli mRNAs? J Mol Biol. 1988 Nov 5;204(1):79–94. doi: 10.1016/0022-2836(88)90601-8. [DOI] [PubMed] [Google Scholar]
- Dunn S. D., Dallmann H. G. An upstream uncD sequence modulates translation of Escherichia coli uncC. J Bacteriol. 1990 May;172(5):2782–2784. doi: 10.1128/jb.172.5.2782-2784.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunn S. D. Effects of the modification of transfer buffer composition and the renaturation of proteins in gels on the recognition of proteins on Western blots by monoclonal antibodies. Anal Biochem. 1986 Aug 15;157(1):144–153. doi: 10.1016/0003-2697(86)90207-1. [DOI] [PubMed] [Google Scholar]
- Dunn S. D., Tozer R. G., Antczak D. F., Heppel L. A. Monoclonal antibodies to Escherichia coli F1-ATPase. Correlation of binding site location with interspecies cross-reactivity and effects on enzyme activity. J Biol Chem. 1985 Sep 5;260(19):10418–10425. [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehretsmann C. P., Carpousis A. J., Krisch H. M. Specificity of Escherichia coli endoribonuclease RNase E: in vivo and in vitro analysis of mutants in a bacteriophage T4 mRNA processing site. Genes Dev. 1992 Jan;6(1):149–159. doi: 10.1101/gad.6.1.149. [DOI] [PubMed] [Google Scholar]
- Foster D. L., Fillingame R. H. Stoichiometry of subunits in the H+-ATPase complex of Escherichia coli. J Biol Chem. 1982 Feb 25;257(4):2009–2015. [PubMed] [Google Scholar]
- Fraker P. J., Speck J. C., Jr Protein and cell membrane iodinations with a sparingly soluble chloroamide, 1,3,4,6-tetrachloro-3a,6a-diphrenylglycoluril. Biochem Biophys Res Commun. 1978 Feb 28;80(4):849–857. doi: 10.1016/0006-291x(78)91322-0. [DOI] [PubMed] [Google Scholar]
- Freier S. M., Kierzek R., Jaeger J. A., Sugimoto N., Caruthers M. H., Neilson T., Turner D. H. Improved free-energy parameters for predictions of RNA duplex stability. Proc Natl Acad Sci U S A. 1986 Dec;83(24):9373–9377. doi: 10.1073/pnas.83.24.9373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibson F., Cox G. B., Downie J. A., Radik J. Partial diploids of Escherichia coli carrying normal and mutant alleles affecting oxidative phosphorylation. Biochem J. 1977 Mar 15;162(3):665–670. doi: 10.1042/bj1620665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Gualerzi C. O., Pon C. L. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990 Jun 26;29(25):5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- Jacques N., Dreyfus M. Translation initiation in Escherichia coli: old and new questions. Mol Microbiol. 1990 Jul;4(7):1063–1067. doi: 10.1111/j.1365-2958.1990.tb00679.x. [DOI] [PubMed] [Google Scholar]
- Kennell D., Riezman H. Transcription and translation initiation frequencies of the Escherichia coli lac operon. J Mol Biol. 1977 Jul;114(1):1–21. doi: 10.1016/0022-2836(77)90279-0. [DOI] [PubMed] [Google Scholar]
- Krieg P. A., Melton D. A. In vitro RNA synthesis with SP6 RNA polymerase. Methods Enzymol. 1987;155:397–415. doi: 10.1016/0076-6879(87)55027-3. [DOI] [PubMed] [Google Scholar]
- Kuki M., Noumi T., Maeda M., Amemura A., Futai M. Functional domains of epsilon subunit of Escherichia coli H+-ATPase (F0F1). J Biol Chem. 1988 Nov 25;263(33):17437–17442. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lesage P., Chiaruttini C., Graffe M., Dondon J., Milet M., Springer M. Messenger RNA secondary structure and translational coupling in the Escherichia coli operon encoding translation initiation factor IF3 and the ribosomal proteins, L35 and L20. J Mol Biol. 1992 Nov 20;228(2):366–386. doi: 10.1016/0022-2836(92)90827-7. [DOI] [PubMed] [Google Scholar]
- Mackie G. A. Secondary structure of the mRNA for ribosomal protein S20. Implications for cleavage by ribonuclease E. J Biol Chem. 1992 Jan 15;267(2):1054–1061. [PubMed] [Google Scholar]
- Mackie G. A. Specific endonucleolytic cleavage of the mRNA for ribosomal protein S20 of Escherichia coli requires the product of the ams gene in vivo and in vitro. J Bacteriol. 1991 Apr;173(8):2488–2497. doi: 10.1128/jb.173.8.2488-2497.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCarthy J. E., Bokelmann C. Determinants of translational initiation efficiency in the atp operon of Escherichia coli. Mol Microbiol. 1988 Jul;2(4):455–465. doi: 10.1111/j.1365-2958.1988.tb00051.x. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990 Mar;6(3):78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- Messing J., Crea R., Seeburg P. H. A system for shotgun DNA sequencing. Nucleic Acids Res. 1981 Jan 24;9(2):309–321. doi: 10.1093/nar/9.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppenheim D. S., Yanofsky C. Translational coupling during expression of the tryptophan operon of Escherichia coli. Genetics. 1980 Aug;95(4):785–795. doi: 10.1093/genetics/95.4.785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patel A. M., Dunn S. D. RNase E-dependent cleavages in the 5' and 3' regions of the Escherichia coli unc mRNA. J Bacteriol. 1992 Jun;174(11):3541–3548. doi: 10.1128/jb.174.11.3541-3548.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy M. K., Apirion D. Purification and properties of ribonuclease E, an RNA-processing enzyme from Escherichia coli. Biochim Biophys Acta. 1983 Sep 28;747(3):200–208. doi: 10.1016/0167-4838(83)90098-5. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer E. M., Hartz D., Gold L., Simoni R. D. Ribosome-binding sites and RNA-processing sites in the transcript of the Escherichia coli unc operon. J Bacteriol. 1989 Jul;171(7):3901–3908. doi: 10.1128/jb.171.7.3901-3908.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T. D., Stormo G. D., Gold L., Ehrenfeucht A. Information content of binding sites on nucleotide sequences. J Mol Biol. 1986 Apr 5;188(3):415–431. doi: 10.1016/0022-2836(86)90165-8. [DOI] [PubMed] [Google Scholar]
- Schulz V. P., Reznikoff W. S. In vitro secondary structure analysis of mRNA from lacZ translation initiation mutants. J Mol Biol. 1990 Jan 20;211(2):427–445. doi: 10.1016/0022-2836(90)90363-Q. [DOI] [PubMed] [Google Scholar]
- Spanjaard R. A., van Duin J. Translational reinitiation in the presence and absence of a Shine and Dalgarno sequence. Nucleic Acids Res. 1989 Jul 25;17(14):5501–5507. doi: 10.1093/nar/17.14.5501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprengel R., Reiss B., Schaller H. Translationally coupled initiation of protein synthesis in Bacillus subtilis. Nucleic Acids Res. 1985 Feb 11;13(3):893–909. doi: 10.1093/nar/13.3.893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L., Ehrenfeucht A. Use of the 'Perceptron' algorithm to distinguish translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2997–3011. doi: 10.1093/nar/10.9.2997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Vincze A., Henderson R. E., McDonald J. J., Leonard N. J. Reaction of diethyl pyrocarbonate with nucleic acid components. Bases and nucleosides derived from guanine, cytosine, and uracil. J Am Chem Soc. 1973 Apr 18;95(8):2677–2682. doi: 10.1021/ja00789a045. [DOI] [PubMed] [Google Scholar]
- Walker J. E., Saraste M., Gay N. J. The unc operon. Nucleotide sequence, regulation and structure of ATP-synthase. Biochim Biophys Acta. 1984 Sep 6;768(2):164–200. doi: 10.1016/0304-4173(84)90003-x. [DOI] [PubMed] [Google Scholar]
- Zuker M., Stiegler P. Optimal computer folding of large RNA sequences using thermodynamics and auxiliary information. Nucleic Acids Res. 1981 Jan 10;9(1):133–148. doi: 10.1093/nar/9.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Control of prokaryotic translational initiation by mRNA secondary structure. Prog Nucleic Acid Res Mol Biol. 1990;38:1–35. doi: 10.1016/s0079-6603(08)60707-2. [DOI] [PubMed] [Google Scholar]