Abstract
Signal transducer and activator of transcription (STAT) proteins have been shown to mediate biological actions in response to cytokines. Stat3, a member of the STAT family, is activated by a variety of cytokines, including the interleukin 6 family of cytokines, leptin, granulocyte colony-stimulating factor, and epidermal growth factor. To address the biological function of Stat3, we generated mice deficient in Stat3 by gene targeting. No viable Stat3-deficient mice could be obtained from heterozygote intercross. Analysis of embryos at several gestation times revealed that Stat3-deficient embryos showed a rapid degeneration between embryonic days 6.5 and 7.5, although they developed into the egg cylinder stage until embryonic day 6.0. These results demonstrate that Stat3 is essential for the early development of mouse embryos.
Signal transducer and activator of transcription (STAT) proteins have been shown to play an important role in cytokine signaling pathways (1, 2). These proteins are tyrosine-phosphorylated by Janus kinases after cytokine binding to its receptor. Once phosphorylated, STAT proteins form homo- or heterodimers, through interaction between the Src homology 2 domain and phosphorylated tyrosine, rapidly translocate to the nucleus and induce several gene expressions. Until now, six members of STAT family, Stat1 through Stat6, have been identified. Each member is shown to be activated by its specific cytokine and responsible for cytokine-mediated responses. Recent studies from mice deficient in several STAT family members have demonstrated that STAT proteins play an essential role in cytokine-mediated biological actions; Stat1 is critical for interferon-mediated actions and innate immunity (3, 4). Stat4 is essential for interleukin (IL)-12-mediated functions and Th1 cell differentiation, whereas Stat6 is for IL-4-mediated functions and Th2 cell differentiation (5–9).
Stat3 was originally identified as acute phase response factor, which is activated by IL-6 family of cytokines (10, 11). This molecule is shown to be important for IL-6-mediated biological effects on cultured cell lines (12, 13). Further studies have demonstrated that Stat3 is activated in response to a variety of cytokines in addition to IL-6 family of cytokines. Stat3 is shown to be tyrosine-phosphorylated by granulocyte colony-stimulating factor and epidermal growth factor (EGF) in cultured cells (11, 14). Furthermore, leptin, a hormone that regulates satiety and energy metabolism, has been shown to induce the activation of Stat3 in the hypothalamus (15). To examine the biological functions of Stat3, we have generated Stat3-deficient mice.
MATERIALS AND METHODS
Generation of Stat3-Deficient Mice.
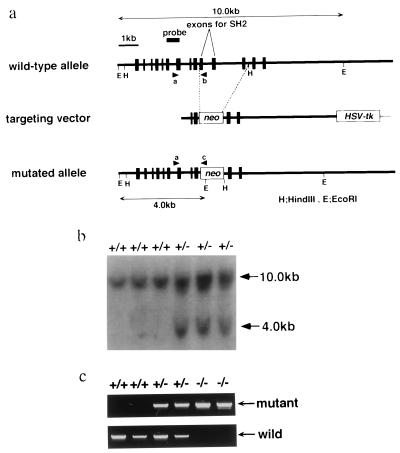
The Stat3 genomic DNA was screened from 129/Sv mouse genomic library, subcloned into pBluescript SK vector (Stratagene), and characterized by restriction enzyme mapping and DNA sequencing as described (16). A targeting vector was designed to replace a 3.0-kb genomic fragment containing exons 20, 21, and 22 with the pMC1-neo (Stratagene). The targeting vector was flanked by the 5.0-kb fragment at 3′ end and the 0.9-kb fragment at 5′ end and contains a HSV-tk cassette at the 3′ end of the vector. The targeting vector was linearized with SalI and electroporated into E14-1 embryonic stem (ES) cells. The resistant clones to G418 and ganciclovir were screened for homologous recombination by PCR and confirmed by Southern blot analysis. Generation of chimeric mice and mutant mice was essentially as described (17). The primers shown in Fig. 1a were: a, 5′-AGCAGCTGACAACGCTGGCTGAGAAGCT-3′; b, 5′-TTGCTGCTCTCGCTGAAGCGCAGTAGG-3′; and c, 5′-ATCGCCTTCTATCGCCTTCTTGACGAG-3′.
Figure 1.
Disruption of the Stat3 gene. (a) The structure of the targeting vector and the mutated Stat3 gene. Restriction sites were: E, EcoRI; H, HindIII. (b) Southern blot analysis of the offspring from intercross of Stat3+/− mice. (c) PCR analysis of the microdissected embryonic tissues at E6.5. Primers a and b were used for detection of the wild-type allele, and a and c were used for the mutated allele.
Histological Analysis.
The decidual swellings were fixed in 4% paraformaldehyde/0.1 M phosphate buffer, pH 7.4, overnight at 4°C, and then immersed in 30% sucrose/0.1 M phosphate buffer for cryoprotection. After 2 days, the tissues were frozen and cut with a cryostat at 16-μm thickness. The tissue sections were stained with hematoxylin and eosin.
PCR Genotyping of Tissue Sections.
Embryonic tissue sections before hematoxylin and eosin staining were microdissected with 26-gauge needle and transferred into 30 μl of lysis buffer (50 mM KCl/1.5 mM MgCl2/10 mM Tris, pH 8.5/0.01% gelatin/0.45% Nonidet P-40/0.45% Tween-20/100 μg/ml proteinase K). Tissues were lysed at 56°C for 1 hr and then incubated at 98°C for 10 min to inactivate proteinase K. Lysis solution (10 μl) was used for templates of PCR amplification.
In Vitro Culture of Blastocyst.
Stat3 heterozygote males and females were intercrossed, and embryonic day 3.5 (E3.5) embryos were collected by flushing from uterus of the plugged females. Blastocysts were independently cultured in 24-well plates coated with 0.1% gelatin in ES medium without leukemia inhibitory factor (LIF). After 5 days of culture, photographs of the cultured embryos were taken, and the sizes of the outgrowths of inner cellular mass were measured. Their genotypes were determined by PCR.
RESULTS
Generation of Stat3-Deficient Mice.
The Stat3 gene was inactivated in ES cells using a targeting vector as shown in Fig. 1a. In this targeting vector, a 3.0-kb fragment of Stat3 genomic DNA including exons 20–22 was replaced with neomycin resistance (neo) gene (Fig. 1a). This deletion causes a loss of Src homology 2 domain and tyrosine residue, which are essential for activation of STAT proteins. ES cells were electroporated with the linearized vector and selected in the presence of G418 and ganciclovir. Two correctly targeted ES cell clones of 300 doubly resistant clones were screened by PCR and confirmed by Southern blot analysis. These ES cell clones were microinjected into C57BL/6 blastocysts to generate chimeric mice, and both successfully contributed to the germ line. Mice heterozygous for the Stat3 mutation (Stat3+/−) were phenotypically normal and fertile. To generate Stat3−/− mice, Stat3+/− mice were intercrossed. The genotypes of offspring were determined at 28 days of age by PCR and Southern blot analysis of tail DNA (Fig. 1b). Of 98 offspring examined, 35 were wild type and 63 were Stat3+/−. No Stat3−/− mice could be detected, indicating homozygous mutation of the Stat3 gene causes embryonic lethality (Table 1).
Table 1.
Genotypes of offspring from Stat3+/− intercross
| Normal phenotypes
|
Abnormal phenotypes | |||
|---|---|---|---|---|
| +/+ | +/− | −/− | ||
| 28 days of age | 35 | 63 | 0 | 0 |
| E14.5 | 2 | 11 | 0 | 6 |
| E11.5 | 4 | 8 | 0 | 5 |
| E9.5 | 3 | 9 | 0 | 4 |
| E8.5 | 5 | 12 | 0 | 8 |
| E7.5 | 4 | 8 | 5 | |
| E7.0 | 3 | 14 | 4 | |
| E6.5 | 4 | 12 | 5 | |
| E6.0 | 4 | 11 | 6 | |
Embryonic Development of Stat3−/− Mice.
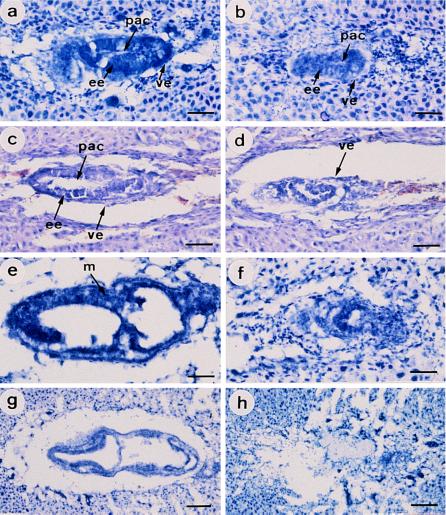
To assess the time of death in utero, embryos from heterozygous intercrosses were dissected and genotyped at several gestation times from E8.5 to E14.5. No Stat3−/− fetuses were identified from decidua containing normal fetuses, and ≈30% of decidua were abnormal with no embryos, indicating that the Stat3−/− mutant concepti die before E8.5 (Table 1). To precisely characterize the differences between wild-type and Stat3−/− embryos, histological analysis were performed at different gestation times from E6.0 to E7.5 (Fig. 2). The genotype was determined by PCR amplification of DNA of the microdissected embryonic samples from tissue sections using primers as indicated in Fig. 1a. Stat3−/− embryos could be detected at the expected Mendelian ratios from E6.0 to E7.5 (Fig. 1c; Table 1). At E6.0, both Stat3−/− and wild-type embryos showed elongation of the egg cylinder, a double-layered structure in which two layers of ectodermal and endodermal cells enclose the proamniotic cavity, although Stat3−/− embryos were smaller than wild-type embryos (Fig. 2 a and b). This finding suggests that Stat3 does not play a critical role in the early postimplantation development, especially in the formation of the egg cylinder. However, by E6.5, Stat3−/− embryos began to degenerate, in contrast to wild-type embryos, which developed to advanced egg cylinder embryos (Fig. 2 c and d). By E7.0, wild-type embryos developed to a primitive streak with the formation of mesoderm between ectoderm and endoderm. In contrast, Stat3−/− embryos were being degenerated with no sign of mesoderm formation (Fig. 2 e and f). By E7.5, all of five Stat3−/− embryos examined were completely resorbed, indicating that Stat3−/− embryos die around E7.0, the day at which gastrulation initiates (Fig. 2 g and h).
Figure 2.
Histological analysis of wild-type and Stat3−/− embryos. Wild-type embryos (a, c, e, and g) and Stat3−/− embryos (b, d, f, and h) were dissected at E6.0 (a and b), E6.5 (c and d), E7.0 (e and f), and E7.5 (g and h). Note the rapid degeneration of Stat3−/− embryos from E6.5 to E7.5. Stat3−/− embryos formed a two-layered egg cylinder at E6.0 (b) but were degenerated rapidly and resorbed completely by E7.5 (d, f, and h). ee, embryonic ectoderm; m, mesoderm; pac, proamniotic cavity; and ve, visceral endoderm. [Bars = 40 μm (a–f) and 100 μm (g and h).]
In Vitro Growth of Stat3−/− Blastocysts.
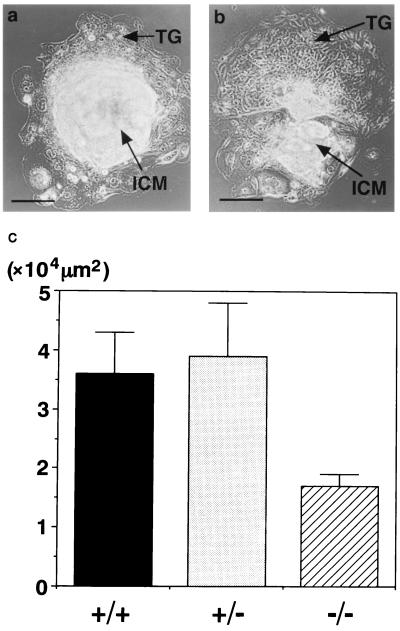
Stat3 is shown to be expressed in ES cells and tyrosine-phosphorylated in response to IL-6 family of cytokines, including LIF. LIF is known to be essential for the maintenance of ES cells in the undifferentiated state. ES cell clones are established from in vitro culture of blastocysts. To directly know the effect of Stat3 deficiency on the growth of blastocysts, E3.5 blastocysts from heterozygous intercross were collected by uterine flushing and cultured in vitro. Stat3−/− blastocyst showed a normal phenotype, indicating that Stat3 deficiency does not affect the development of preimplantation period. After 5 days of culture, both Stat3−/− and wild-type blastocysts produced trophoblast giant cell outgrowth. Furthermore, the inner cellular mass (ICM) of Stat3−/− blastocyst did show the outgrowth, although the size of outgrowth was smaller than that of wild type (Fig. 3). These results indicate that Stat3 deficiency does not critically affect the outgrowth of blastocysts in spite of the fact that it causes the early embryonic lethality.
Figure 3.
In vitro outgrowth of blastocysts. Blastocysts were cultured for 5 days, then photographed, lysed, and PCR-genotyped. (a) In vitro cultured wild-type blastocyst displaying outgrowth of trophoblast giant cells and ICM. (b) Cultured Stat3−/− blastocyst also displaying outgrowth of trophoblast giant cells and ICM, but the size of the ICM outgrowth was smaller than that of wild type. TG, trophoblast giant cell; ICM, inner cellular mass. (Bar = 40 μm.) (c) Graph of ICM outgrowth size. The shapes of ICM outgrowth were approximately ellipses, and surface areas were measured after 5 days of culture.
DISCUSSION
This study demonstrates the functional importance of Stat3 in early embryonic development of the mouse. Stat3−/− embryos developed to the egg cylinder-stage embryos like wild-type embryos until E6.0. This was confirmed by in vitro culture experiment of Stat3−/− blastocysts, which displayed the outgrowth of ICM. But the sizes of E6.0 Stat3−/− embryos and the outgrowth of ICM of Stat3−/− blastocysts were smaller than those of wild type. These findings indicate that Stat3 is not essential for the formation of the egg cylinder, but in some extent it is responsible for the cell growth in this period. Stat3−/− embryos developed until E6.0, however, rapidly degenerated with no obvious mesoderm formation between E6.5 and E7.5. Our preliminary data from in situ hybridization analysis of embryos between E5.5 and E7.5 showed that Stat3 mRNA began to be expressed exclusively in visceral endoderm around E6.0. In addition to embryonic expression, high levels of Stat3 expression were detected in the decidua. Visceral endoderm, which covers the upper side of the egg cylinder embryos, is known to have an important function in mediating metabolic exchange with maternal blood (18). The onset of degeneration of Stat3−/− embryos coincided with the beginning of Stat3 expression in visceral endoderm. Therefore, we speculate that the lethality may be due to the result of a defect in functions of visceral endoderm, such as nutritional insufficiency.
Stat3 can be activated in response to a number of different cytokines and growth factors, including the IL-6 family of cytokines [IL-6, IL-11, LIF, ciliary neurotrophic factor (CNTF), and oncostatin M], granulocyte colony-stimulating factor, and leptin, all of which use glycoprotein 130 (gp130) or closely related signaling molecules, such as LIF receptor (LIF-R), granulocyte colony-stimulating factor receptor, and leptin receptor (10, 11, 14, 15). Stat3 is also activated in response to EGF (11). These receptor molecules harbor a common Stat3 docking motif (YXXQ) in the cytoplasmic domain. Among target inactivation of the cytokines and receptor molecules that have been shown to activate Stat3, mice deficient in gp130, LIF-R, CNTF-R, and EGF-R are lethal. However, the period of embryonic death observed with the Stat3−/− mice is much earlier: LIF-R−/− mice and CNTF-R−/− mice die shortly after birth (19–21). gp130−/− mice die around E15.5–18.5 as a result of abnormal heart, placenta, and hematopoiesis (22). The death of EGF-R−/− mice was observed after E11.5 on a 129/Sv × C57BL/6 background (23, 24). Therefore, the early embryonic lethality in Stat3−/− mice may be due to loss of a combined effect of two or more of these receptor-mediated signals. Alternatively, Stat3 may be activated through an as yet unidentified signaling molecule critical for early embryonic development. Furthermore, our present study clearly shows that Stat3 plays a unique role in early embryonic development, which cannot be compensated by other STAT family members despite a wide utilization of Stat1, Stat3, and Stat5 by a variety of cytokine receptors. The conditional knockout study of the Stat3 gene using Cre-loxP system will delineate the cytokines that use Stat3 for their biological actions in later developmental stages and in adult mice.
Acknowledgments
We thank Y. Kataoka, M. Inoue, F. Ueno, and K. Ishikawa for technical assistance, K. Miki for histological analysis, and K. Kubota, K. Ogishi, and T. Aoki for secretarial assistance. This work was supported by grants from the Ministry of Education of Japan.
ABBREVIATIONS
- STAT
signal transducer and activator of transcription
- IL
interleukin
- ES
embryonic stem
- E
embryonic day
- LIF
leukemia inhibitory factor
- ICM
inner cellular mass
- EGF
epidermal growth factor
- R
receptor
References
- 1.Schindler C, Darnell J E., Jr Annu Rev Biochem. 1995;64:621–651. doi: 10.1146/annurev.bi.64.070195.003201. [DOI] [PubMed] [Google Scholar]
- 2.Ihle J N. Cell. 1996;84:331–334. doi: 10.1016/s0092-8674(00)81277-5. [DOI] [PubMed] [Google Scholar]
- 3.Meraz M A, White J M, Sheehan K C F, Bach E A, Rodig S J, Dighe A S, Kaplan D H, Riley J K, Greenlund A C, Campbell D, Carver-Moore K, DuBois R N, Clark R, Aguet M, Schreiber R D. Cell. 1996;84:431–442. doi: 10.1016/s0092-8674(00)81288-x. [DOI] [PubMed] [Google Scholar]
- 4.Durbin J E, Hackenmiller R, Simon M C, Levy D E. Cell. 1996;84:443–450. doi: 10.1016/s0092-8674(00)81289-1. [DOI] [PubMed] [Google Scholar]
- 5.Takeda K, Tanaka T, Shi W, Matsumoto M, Minami M, Kashiwamura S, Nakanishi K, Yoshida N, Kishimoto T, Akira S. Nature (London) 1996;380:627–630. doi: 10.1038/380627a0. [DOI] [PubMed] [Google Scholar]
- 6.Shimoda K, Deursen J, Sangster M Y, Sarawar S R, Carson R T, Tripp R A, Chu C, Quelle F W, Nosaka T, Vignali D A A, Doherty P C, Grosveld G, Paul W E, Ihle J N. Nature (London) 1996;380:630–633. doi: 10.1038/380630a0. [DOI] [PubMed] [Google Scholar]
- 7.Kaplan M H, Schindler U, Smiley S T, Grusby M J. Immunity. 1996;4:313–319. doi: 10.1016/s1074-7613(00)80439-2. [DOI] [PubMed] [Google Scholar]
- 8.Thierfelder W E, Deursen J, Yamamoto K, Tripp R A, Sarawar S R, Carson R T, Sangster M Y, Vignali D A A, Doherty P C, Grosveld G C, Ihle J N. Nature (London) 1996;382:171–174. doi: 10.1038/382171a0. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan M H, Sun Y-L, Hoey T, Grusby M J. Nature (London) 1996;382:174–177. doi: 10.1038/382174a0. [DOI] [PubMed] [Google Scholar]
- 10.Akira S, Nishio Y, Inoue M, Wang X J, Wei S, Matsusaka T, Yoshida K, Sudo T, Naruto M, Kishimoto T. Cell. 1994;77:63–71. doi: 10.1016/0092-8674(94)90235-6. [DOI] [PubMed] [Google Scholar]
- 11.Zhong Z, Wen Z, Darnell J E., Jr Science. 1994;264:95–98. doi: 10.1126/science.8140422. [DOI] [PubMed] [Google Scholar]
- 12.Minami M, Inoue M, Wei S, Takeda K, Matsumoto M, Kishimoto T, Akira S. Proc Natl Acad Sci USA. 1996;93:3963–3966. doi: 10.1073/pnas.93.9.3963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yamanaka Y, Nakajima K, Fukada T, Hibi M, Hirano T. EMBO J. 1996;15:1557–1565. [PMC free article] [PubMed] [Google Scholar]
- 14.Tian S-S, Lamb P, Seidel H M, Stein R B, Rosen J. Blood. 1994;84:1760–1764. [PubMed] [Google Scholar]
- 15.Vaisse C, Halaas J L, Horvath C M, Darnell J E, Jr, Stoffel M, Friedman J M. Nat Genet. 1996;14:95–97. doi: 10.1038/ng0996-95. [DOI] [PubMed] [Google Scholar]
- 16.Shi W, Inoue M, Minami M, Takeda K, Matsumoto M, Matsuda Y, Kishimoto T, Akira S. Int Immunol. 1996;8:1205–1211. doi: 10.1093/intimm/8.8.1205. [DOI] [PubMed] [Google Scholar]
- 17.Tanaka T, Akira S, Yoshida K, Umemoto M, Yoneda Y, Shirafuji N, Fujiwara H, Suematsu S, Yoshida N, Kishimoto T. Cell. 1995;80:353–361. doi: 10.1016/0092-8674(95)90418-2. [DOI] [PubMed] [Google Scholar]
- 18.Cross J C, Werb Z, Fisher S J. Science. 1994;266:1508–1518. doi: 10.1126/science.7985020. [DOI] [PubMed] [Google Scholar]
- 19.Ware C B, Horowitz M C, Renshaw B R, Hunt J S, Liggitt D, Koblar S A, Gliniak B C, McKenna H J, Papayannopoulou T, Thoma B, Cheng L, Donovan P J, Peschon J J, Bartlett P F, Willis C R, Wright B D, Carpenter M K, Davidson B L, Gearing D P. Development (Cambridge, UK) 1995;121:1283–1299. doi: 10.1242/dev.121.5.1283. [DOI] [PubMed] [Google Scholar]
- 20.Li M, Sendtner M, Smith A. Nature (London) 1995;378:724–727. doi: 10.1038/378724a0. [DOI] [PubMed] [Google Scholar]
- 21.DeChiara T M, Vejsada R, Poueymirou W T, Acheson A, Suri C, Conover J C, Friedman B, McClain J, Pan L, Stahl N, Ip N Y, Kato A, Yancopoulos G D. Cell. 1995;83:313–322. doi: 10.1016/0092-8674(95)90172-8. [DOI] [PubMed] [Google Scholar]
- 22.Yoshida K, Taga T, Saito M, Suematsu S, Kumanogoh A, Tanaka T, Fujiwara H, Hirata M, Yamagami T, Nakahata T, Hirabayashi T, Yoneda Y, Tanaka K, Wang W-Z, Mori C, Shiota K, Yoshida N, Kishimoto T. Proc Natl Acad Sci USA. 1996;93:407–411. doi: 10.1073/pnas.93.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sibilia M, Wagner E F. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- 24.Miettinen P, Berger J E, Meneses J, Phung Y, Pedersen R A, Werb Z, Derynck R. Nature (London) 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]