Abstract
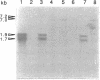
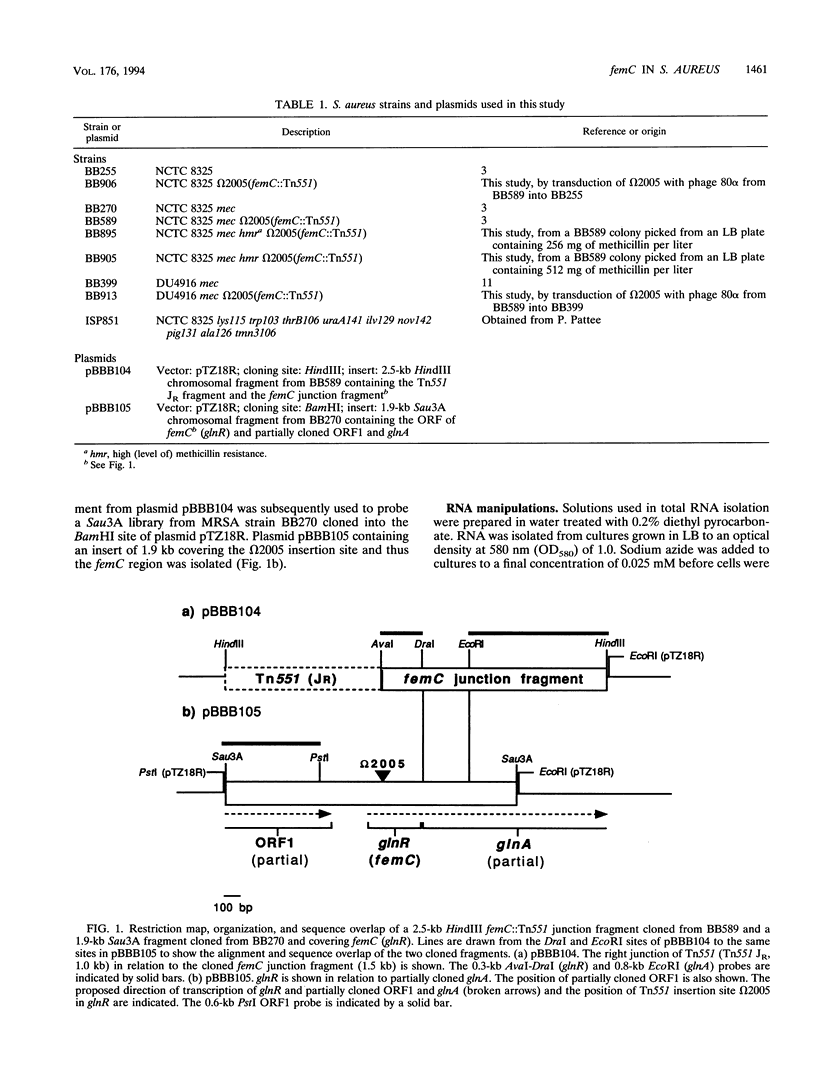
Tn551 insertional inactivation of femC is known to reduce methicillin resistance levels in methicillin-resistant and -susceptible Staphylococcus aureus. By use of cotransductional crosses, femC was mapped close to thrB on the SmaI-A fragment of the S. aureus NCTC 8325 chromosome. The Tn551 insertion femC::omega 2005 was found to interrupt an open reading frame coding for a putative protein of 121 amino acids which is highly similar to the glutamine synthetase repressors (GlnR) of Bacillus spp. Downstream of femC, an open reading frame highly similar to Bacillus sp. glutamine synthetases (GlnA) was found. Northern (RNA) blots probed with putative glnR or glnA fragments revealed that 1.7- and 1.9-kb transcripts characteristic of wild-type cells were replaced by less abundant 7.0- and 7.2-kb transcripts in the femC::omega 2005 mutant. Total glutamine synthetase activity was also decreased in the mutant strain; the addition of glutamine to defined media restored the wild-type methicillin resistance phenotype of the femC mutant. This result suggests that the omega 2005 insertion in glnR has a polar effect on glnA and that glnR and glnA are transcribed together as an operon. These results suggest that the loss of wild-type levels of glutamine synthetase and the consequent decrease in glutamine availability cause a decreased level of methicillin resistance.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Berger-Bächi B., Barberis-Maino L., Strässle A., Kayser F. H. FemA, a host-mediated factor essential for methicillin resistance in Staphylococcus aureus: molecular cloning and characterization. Mol Gen Genet. 1989 Oct;219(1-2):263–269. doi: 10.1007/BF00261186. [DOI] [PubMed] [Google Scholar]
- Berger-Bächi B. Insertional inactivation of staphylococcal methicillin resistance by Tn551. J Bacteriol. 1983 Apr;154(1):479–487. doi: 10.1128/jb.154.1.479-487.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berger-Bächi B., Strässle A., Gustafson J. E., Kayser F. H. Mapping and characterization of multiple chromosomal factors involved in methicillin resistance in Staphylococcus aureus. Antimicrob Agents Chemother. 1992 Jul;36(7):1367–1373. doi: 10.1128/aac.36.7.1367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambers H. F. Methicillin-resistant staphylococci. Clin Microbiol Rev. 1988 Apr;1(2):173–186. doi: 10.1128/cmr.1.2.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deuel T. F., Stadtman E. R. Some kinetic properties of Bacillus subtilis glutamine synthetase. J Biol Chem. 1970 Oct 25;245(20):5206–5213. [PubMed] [Google Scholar]
- Fisher S. H., Sonenshein A. L. Bacillus subtilis glutamine synthetase mutants pleiotropically altered in glucose catabolite repression. J Bacteriol. 1984 Feb;157(2):612–621. doi: 10.1128/jb.157.2.612-621.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafson J. E., Berger-Bächi B., Strässle A., Wilkinson B. J. Autolysis of methicillin-resistant and -susceptible Staphylococcus aureus. Antimicrob Agents Chemother. 1992 Mar;36(3):566–572. doi: 10.1128/aac.36.3.566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henze U., Sidow T., Wecke J., Labischinski H., Berger-Bächi B. Influence of femB on methicillin resistance and peptidoglycan metabolism in Staphylococcus aureus. J Bacteriol. 1993 Mar;175(6):1612–1620. doi: 10.1128/jb.175.6.1612-1620.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hächler H., Berger-Bächi B., Kayser F. H. Genetic characterization of a Clostridium difficile erythromycin-clindamycin resistance determinant that is transferable to Staphylococcus aureus. Antimicrob Agents Chemother. 1987 Jul;31(7):1039–1045. doi: 10.1128/aac.31.7.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishino Y., Morgenthaler P., Hottinger H., Söll D. Organization and nucleotide sequence of the glutamine synthetase (glnA) gene from Lactobacillus delbrueckii subsp. bulgaricus. Appl Environ Microbiol. 1992 Sep;58(9):3165–3169. doi: 10.1128/aem.58.9.3165-3169.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum J., Hartman B. J., Novick R. P., Tomasz A. Conversion of a homogeneously methicillin-resistant strain of Staphylococcus aureus to heterogeneous resistance by Tn551-mediated insertional inactivation. Eur J Clin Microbiol. 1986 Dec;5(6):714–718. doi: 10.1007/BF02013311. [DOI] [PubMed] [Google Scholar]
- Kumada Y., Benson D. R., Hillemann D., Hosted T. J., Rochefort D. A., Thompson C. J., Wohlleben W., Tateno Y. Evolution of the glutamine synthetase gene, one of the oldest existing and functioning genes. Proc Natl Acad Sci U S A. 1993 Apr 1;90(7):3009–3013. doi: 10.1073/pnas.90.7.3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labischinski H. Consequences of the interaction of beta-lactam antibiotics with penicillin binding proteins from sensitive and resistant Staphylococcus aureus strains. Med Microbiol Immunol. 1992;181(5):241–265. doi: 10.1007/BF00198846. [DOI] [PubMed] [Google Scholar]
- Maidhof H., Reinicke B., Blümel P., Berger-Bächi B., Labischinski H. femA, which encodes a factor essential for expression of methicillin resistance, affects glycine content of peptidoglycan in methicillin-resistant and methicillin-susceptible Staphylococcus aureus strains. J Bacteriol. 1991 Jun;173(11):3507–3513. doi: 10.1128/jb.173.11.3507-3513.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano Y., Kato C., Tanaka E., Kimura K., Horikoshi K. Nucleotide sequence of the glutamine synthetase gene (glnA) and its upstream region from Bacillus cereus. J Biochem. 1989 Aug;106(2):209–215. doi: 10.1093/oxfordjournals.jbchem.a122834. [DOI] [PubMed] [Google Scholar]
- Nakel M., Ghuysen J. M., Kandler O. Wall peptidoglycan in Aerococcus viridans strains 201 Evans and ATCC 11563 and in Gaffkya homari strain ATCC 10400. Biochemistry. 1971 May 25;10(11):2170–2175. doi: 10.1021/bi00787a033. [DOI] [PubMed] [Google Scholar]
- SUTHERLAND R., ROLINSON G. N. CHARACTERISTICS OF METHICILLIN-RESISTANT STAPHYLOCOCCI. J Bacteriol. 1964 Apr;87:887–899. doi: 10.1128/jb.87.4.887-899.1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreier H. J., Sonenshein A. L. Altered regulation of the glnA gene in glutamine synthetase mutants of Bacillus subtilis. J Bacteriol. 1986 Jul;167(1):35–43. doi: 10.1128/jb.167.1.35-43.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siewert G., Strominger J. L. Biosynthesis of the peptidoglycan of bacterial cell walls. XI. Formation of the isoglutamine amide group in the cell walls of Staphylococcus aureus. J Biol Chem. 1968 Feb 25;243(4):783–790. [PubMed] [Google Scholar]
- Strauch M. A., Aronson A. I., Brown S. W., Schreier H. J., Sonenhein A. L. Sequence of the Bacillus subtilis glutamine synthetase gene region. Gene. 1988 Nov 30;71(2):257–265. doi: 10.1016/0378-1119(88)90042-x. [DOI] [PubMed] [Google Scholar]
- Tipper D. J., Katz W., Strominger J. L., Ghuysen J. M. Substituents on the alpha-carboxyl group of D-glutamic acid in the peptidoglycan of several bacterial cell walls. Biochemistry. 1967 Mar;6(3):921–929. doi: 10.1021/bi00855a036. [DOI] [PubMed] [Google Scholar]
- Tipper D. J., Strominger J. L. Mechanism of action of penicillins: a proposal based on their structural similarity to acyl-D-alanyl-D-alanine. Proc Natl Acad Sci U S A. 1965 Oct;54(4):1133–1141. doi: 10.1073/pnas.54.4.1133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Townsend D. E., Wilkinson B. J. Proline transport in Staphylococcus aureus: a high-affinity system and a low-affinity system involved in osmoregulation. J Bacteriol. 1992 Apr;174(8):2702–2710. doi: 10.1128/jb.174.8.2702-2710.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woese C. R. Bacterial evolution. Microbiol Rev. 1987 Jun;51(2):221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woolfolk C. A., Shapiro B., Stadtman E. R. Regulation of glutamine synthetase. I. Purification and properties of glutamine synthetase from Escherichia coli. Arch Biochem Biophys. 1966 Sep 26;116(1):177–192. doi: 10.1016/0003-9861(66)90026-9. [DOI] [PubMed] [Google Scholar]
- de Jonge B. L., Chang Y. S., Gage D., Tomasz A. Peptidoglycan composition in heterogeneous Tn551 mutants of a methicillin-resistant Staphylococcus aureus strain. J Biol Chem. 1992 Jun 5;267(16):11255–11259. [PubMed] [Google Scholar]
- de Jonge B. L., Chang Y. S., Gage D., Tomasz A. Peptidoglycan composition of a highly methicillin-resistant Staphylococcus aureus strain. The role of penicillin binding protein 2A. J Biol Chem. 1992 Jun 5;267(16):11248–11254. [PubMed] [Google Scholar]