Abstract
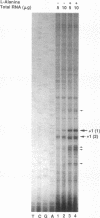
A fragment of the Escherichia coli K-12 chromosome complementing the D-amino acid dehydrogenase and catabolic alanine racemase deficiency of a dad operon deletion mutant was cloned in a mini-Mu plasmid. The dadA and dadX genes were localized to a 3.5-kb part of the plasmid insert. The nucleotide sequence of this fragment revealed two open reading frames encoding 432- and 356-amino-acid-long proteins. We show here that they correspond to the dadA and dadX genes. The dadA gene can encode only the smaller of the two subunits of D-amino acid dehydrogenase. A computer search revealed the presence of a flavin adenine dinucleotide-binding motif in the N-terminal domain of the deduced DadA protein sequence. This is in agreement with biochemical data showing that the D-amino acid dehydrogenase contains flavin adenine dinucleotide in its active center. The predicted dadX gene product appeared to be 85% identical to a dadB-encoded catabolic alanine racemase of Salmonella typhimurium. The organization of the dadA and dadX genes confirmed our previous conclusion based on the genetic data (J. Wild, J. Hennig, M. Lobocka, W. Walczak, and T. Kłopotowski, Mol. Gen. Genet. 198:315-322, 1985) that these genes form an operon. The main transcription start points of the dad operon were determined by primer extension. They are preceded by a putative sigma 70 promoter sequence and two cyclic AMP-cyclic AMP receptor protein (cAMP-CRP) binding sites, one of higher and one of lower affinity to CRP. We propose that the high-affinity site, centered 59.5 bp upstream of the main transcription start point, plays a role in cAMP-CRP-mediated activation of dad operon expression in the absence of glucose.
Full text
PDF









Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Hanamura A., Tobe T. Semisynthetic promoters activated by cyclic AMP receptor protein of Escherichia coli. Gene. 1989 Dec 21;85(1):91–97. doi: 10.1016/0378-1119(89)90468-x. [DOI] [PubMed] [Google Scholar]
- Badet B., Roise D., Walsh C. T. Inactivation of the dadB Salmonella typhimurium alanine racemase by D and L isomers of beta-substituted alanines: kinetics, stoichiometry, active site peptide sequencing, and reaction mechanism. Biochemistry. 1984 Oct 23;23(22):5188–5194. doi: 10.1021/bi00317a016. [DOI] [PubMed] [Google Scholar]
- Beelen R. H., Feldmann A. M., Wijsman H. J. A regulatory gene and a structural gene for alaninase in Escherichia coli. Mol Gen Genet. 1973 Mar 19;121(4):369–374. doi: 10.1007/BF00433235. [DOI] [PubMed] [Google Scholar]
- Belfort M., Pedersen-Lane J., West D., Ehrenman K., Maley G., Chu F., Maley F. Processing of the intron-containing thymidylate synthase (td) gene of phage T4 is at the RNA level. Cell. 1985 Jun;41(2):375–382. doi: 10.1016/s0092-8674(85)80010-6. [DOI] [PubMed] [Google Scholar]
- Bochner B. R., Savageau M. A. Generalized indicator plate for genetic, metabolic, and taxonomic studies with microorganisms. Appl Environ Microbiol. 1977 Feb;33(2):434–444. doi: 10.1128/aem.33.2.434-444.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Botsford J. L., Harman J. G. Cyclic AMP in prokaryotes. Microbiol Rev. 1992 Mar;56(1):100–122. doi: 10.1128/mr.56.1.100-122.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracco L., Kotlarz D., Kolb A., Diekmann S., Buc H. Synthetic curved DNA sequences can act as transcriptional activators in Escherichia coli. EMBO J. 1989 Dec 20;8(13):4289–4296. doi: 10.1002/j.1460-2075.1989.tb08615.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brendel V., Trifonov E. N. A computer algorithm for testing potential prokaryotic terminators. Nucleic Acids Res. 1984 May 25;12(10):4411–4427. doi: 10.1093/nar/12.10.4411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Ebright Y. W., Gunasekera A. Consensus DNA site for the Escherichia coli catabolite gene activator protein (CAP): CAP exhibits a 450-fold higher affinity for the consensus DNA site than for the E. coli lac DNA site. Nucleic Acids Res. 1989 Dec 25;17(24):10295–10305. doi: 10.1093/nar/17.24.10295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin F. C., Venables W. A. Biochemical, genetic, and regulatory studies of alanine catabolism in Escherichia coli K12. Mol Gen Genet. 1976 Dec 8;149(2):229–237. doi: 10.1007/BF00332894. [DOI] [PubMed] [Google Scholar]
- Franklin F. C., Venables W. A., Wijsman H. J. Genetic studies of D-alanine-dehydrogenase-less mutants of Escherichia coli K12. Genet Res. 1981 Oct;38(2):197–208. doi: 10.1017/s0016672300020528. [DOI] [PubMed] [Google Scholar]
- Fukui K., Watanabe F., Shibata T., Miyake Y. Molecular cloning and sequence analysis of cDNAs encoding porcine kidney D-amino acid oxidase. Biochemistry. 1987 Jun 16;26(12):3612–3618. doi: 10.1021/bi00386a054. [DOI] [PubMed] [Google Scholar]
- Galakatos N. G., Daub E., Botstein D., Walsh C. T. Biosynthetic alr alanine racemase from Salmonella typhimurium: DNA and protein sequence determination. Biochemistry. 1986 Jun 3;25(11):3255–3260. doi: 10.1021/bi00359a026. [DOI] [PubMed] [Google Scholar]
- Galakatos N. G., Walsh C. T. Specific proteolysis of native alanine racemases from Salmonella typhimurium: identification of the cleavage site and characterization of the clipped two-domain proteins. Biochemistry. 1987 Dec 15;26(25):8475–8480. doi: 10.1021/bi00399a066. [DOI] [PubMed] [Google Scholar]
- Gartenberg M. R., Crothers D. M. DNA sequence determinants of CAP-induced bending and protein binding affinity. Nature. 1988 Jun 30;333(6176):824–829. doi: 10.1038/333824a0. [DOI] [PubMed] [Google Scholar]
- Gaston K., Bell A., Kolb A., Buc H., Busby S. Stringent spacing requirements for transcription activation by CRP. Cell. 1990 Aug 24;62(4):733–743. doi: 10.1016/0092-8674(90)90118-x. [DOI] [PubMed] [Google Scholar]
- Groisman E. A., Casadaban M. J. Mini-mu bacteriophage with plasmid replicons for in vivo cloning and lac gene fusing. J Bacteriol. 1986 Oct;168(1):357–364. doi: 10.1128/jb.168.1.357-364.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi H., Wada H., Yoshimura T., Esaki N., Soda K. Recent topics in pyridoxal 5'-phosphate enzyme studies. Annu Rev Biochem. 1990;59:87–110. doi: 10.1146/annurev.bi.59.070190.000511. [DOI] [PubMed] [Google Scholar]
- Hinton D. M. Transcript analyses of the uvsX-40-41 region of bacteriophage T4. Changes in the RNA as infection proceeds. J Biol Chem. 1989 Aug 25;264(24):14432–14439. [PubMed] [Google Scholar]
- Irani M., Musso R., Adhya S. Cyclic-AMP-dependent switch in initiation of transcription from the two promoters of the Escherichia coli gal operon: identification and assay of 5'-triphosphate ends of mRNA by GTP:RNA guanyltransferase. J Bacteriol. 1989 Mar;171(3):1623–1630. doi: 10.1128/jb.171.3.1623-1630.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isogai T., Ono H., Ishitani Y., Kojo H., Ueda Y., Kohsaka M. Structure and expression of cDNA for D-amino acid oxidase active against cephalosporin C from Fusarium solani. J Biochem. 1990 Dec;108(6):1063–1069. doi: 10.1093/oxfordjournals.jbchem.a123306. [DOI] [PubMed] [Google Scholar]
- Jones H., Venables W. A. Effects of solubilisation on some properties of the membrane-bound respiratory enzyme D-amino acid dehydrogenase of Escherichia coli. FEBS Lett. 1983 Jan 24;151(2):189–192. doi: 10.1016/0014-5793(83)80066-0. [DOI] [PubMed] [Google Scholar]
- Jones H., Venables W. A. Solubilisation of D-amino acid dehydrogenase of Escherichia coli K12 and its re-binding to envelope preparations. Biochimie. 1983 Mar;65(3):177–183. doi: 10.1016/s0300-9084(83)80082-0. [DOI] [PubMed] [Google Scholar]
- Kaczorowski G., Shaw L., F-entes M., Walsh C. Coupling of alanine racemase and D-alanine dehydrogenase to active transport of amino acids in Escherichia coli B membrane vesicles. J Biol Chem. 1975 Apr 25;250(8):2855–2865. [PubMed] [Google Scholar]
- Kaczorowski G., Walsh C. Active transport in Excherichia coli B membrane vesicles. Irreversible uncoupling by chloropyruvate. J Biol Chem. 1975 Dec 10;250(23):8931–8937. [PubMed] [Google Scholar]
- Kieny M. P., Lathe R., Lecocq J. P. New versatile cloning and sequencing vectors based on bacteriophage M13. Gene. 1983 Dec;26(1):91–99. doi: 10.1016/0378-1119(83)90039-2. [DOI] [PubMed] [Google Scholar]
- Komeda Y., Iino T. Regulation of expression of the flagellin gene (hag) in Escherichia coli K-12: analysis of hag-lac gene fusions. J Bacteriol. 1979 Sep;139(3):721–729. doi: 10.1128/jb.139.3.721-729.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lambert M. P., Neuhaus F. C. Factors affecting the level of alanine racemase in Escherichia coli. J Bacteriol. 1972 Mar;109(3):1156–1161. doi: 10.1128/jb.109.3.1156-1161.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Momoi K., Fukui K., Tada M., Miyake Y. Gene expression of D-amino acid oxidase in rabbit kidney. J Biochem. 1990 Sep;108(3):406–413. doi: 10.1093/oxfordjournals.jbchem.a123214. [DOI] [PubMed] [Google Scholar]
- Momoi K., Fukui K., Watanabe F., Miyake Y. Molecular cloning and sequence analysis of cDNA encoding human kidney D-amino acid oxidase. FEBS Lett. 1988 Sep 26;238(1):180–184. doi: 10.1016/0014-5793(88)80252-7. [DOI] [PubMed] [Google Scholar]
- Mulligan M. E., Hawley D. K., Entriken R., McClure W. R. Escherichia coli promoter sequences predict in vitro RNA polymerase selectivity. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):789–800. doi: 10.1093/nar/12.1part2.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsiewski P. J., Kaczorowski G. J., Walsh C. T., Kaback H. R. Reconstitution of Escherichia coli membrane vesicles with D-amino acid dehydrogenase. Biochemistry. 1981 Oct 13;20(21):6272–6279. doi: 10.1021/bi00524a056. [DOI] [PubMed] [Google Scholar]
- Olsiewski P. J., Kaczorowski G. J., Walsh C. Purification and properties of D-amino acid dehydrogenase, an inducible membrane-bound iron-sulfur flavoenzyme from Escherichia coli B. J Biol Chem. 1980 May 25;255(10):4487–4494. [PubMed] [Google Scholar]
- Rodier F., Sallantin J. Localization of the initiation of translation in messenger RNAs of prokaryotes by learning techniques. Biochimie. 1985 May;67(5):533–539. doi: 10.1016/s0300-9084(85)80273-x. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz A. Cyclic AMP receptor proteins interacts with lactose operator DNA. Nucleic Acids Res. 1981 Jan 24;9(2):277–292. doi: 10.1093/nar/9.2.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shine J., Dalgarno L. The 3'-terminal sequence of Escherichia coli 16S ribosomal RNA: complementarity to nonsense triplets and ribosome binding sites. Proc Natl Acad Sci U S A. 1974 Apr;71(4):1342–1346. doi: 10.1073/pnas.71.4.1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simons R. W., Egan P. A., Chute H. T., Nunn W. D. Regulation of fatty acid degradation in Escherichia coli: isolation and characterization of strains bearing insertion and temperature-sensitive mutations in gene fadR. J Bacteriol. 1980 May;142(2):621–632. doi: 10.1128/jb.142.2.621-632.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson R. B. Interaction of the cAMP receptor protein with the lac promoter. Nucleic Acids Res. 1980 Feb 25;8(4):759–766. [PMC free article] [PubMed] [Google Scholar]
- Tada M., Fukui K., Momoi K., Miyake Y. Cloning and expression of a cDNA encoding mouse kidney D-amino acid oxidase. Gene. 1990 Jun 15;90(2):293–297. doi: 10.1016/0378-1119(90)90193-u. [DOI] [PubMed] [Google Scholar]
- Tanizawa K., Ohshima A., Scheidegger A., Inagaki K., Tanaka H., Soda K. Thermostable alanine racemase from Bacillus stearothermophilus: DNA and protein sequence determination and secondary structure prediction. Biochemistry. 1988 Feb 23;27(4):1311–1316. doi: 10.1021/bi00404a033. [DOI] [PubMed] [Google Scholar]
- Thornberry N. A., Bull H. G., Taub D., Wilson K. E., Giménez-Gallego G., Rosegay A., Soderman D. D., Patchett A. A. Mechanism-based inactivation of alanine racemase by 3-halovinylglycines. J Biol Chem. 1991 Nov 15;266(32):21657–21665. [PubMed] [Google Scholar]
- Walsh C. T. Enzymes in the D-alanine branch of bacterial cell wall peptidoglycan assembly. J Biol Chem. 1989 Feb 15;264(5):2393–2396. [PubMed] [Google Scholar]
- Wasserman S. A., Daub E., Grisafi P., Botstein D., Walsh C. T. Catabolic alanine racemase from Salmonella typhimurium: DNA sequence, enzyme purification, and characterization. Biochemistry. 1984 Oct 23;23(22):5182–5187. doi: 10.1021/bi00317a015. [DOI] [PubMed] [Google Scholar]
- Wasserman S. A., Walsh C. T., Botstein D. Two alanine racemase genes in Salmonella typhimurium that differ in structure and function. J Bacteriol. 1983 Mar;153(3):1439–1450. doi: 10.1128/jb.153.3.1439-1450.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wild J., Filutowicz M., Kłopotowski T. Utilization of D-amino acids by dadR mutants of Salmonella typhimurium. Arch Microbiol. 1978 Jul;118(1):71–77. doi: 10.1007/BF00406077. [DOI] [PubMed] [Google Scholar]
- Wild J., Hennig J., Lobocka M., Walczak W., Kłopotowski T. Identification of the dadX gene coding for the predominant isozyme of alanine racemase in Escherichia coli K12. Mol Gen Genet. 1985;198(2):315–322. doi: 10.1007/BF00383013. [DOI] [PubMed] [Google Scholar]
- Wild J., Klopotowski T. D-Amino acid dehydrogenase of Escherichia coli K12: positive selection of mutants defective in enzyme activity and localization of the structural gene. Mol Gen Genet. 1981;181(3):373–378. doi: 10.1007/BF00425614. [DOI] [PubMed] [Google Scholar]
- Wild J., Kłopotowski T. Insensitivity of D-amino acid dehydrogenase synthesis to catabolic repression in dadR mutants of Salmonella typhimurium. Mol Gen Genet. 1975;136(1):63–73. doi: 10.1007/BF00275449. [DOI] [PubMed] [Google Scholar]
- Wild J., Obrepalska B. Regulation of expression of the dadA gene encoding D-amino acid dehydrogenase in Escherichia coli: analysis of dadA-lac fusions and direction of dadA transcription. Mol Gen Genet. 1982;186(3):405–410. doi: 10.1007/BF00729461. [DOI] [PubMed] [Google Scholar]
- Wild J., Zakrzewska B., Walczak W., Kłopotowski T. Two distinct types of mutations conferring to Escherichia coli K12 capability of D-tryptophan utilization. Acta Microbiol Pol. 1987;36(1-2):17–28. [PubMed] [Google Scholar]