Abstract
Freezing avoidance conferred by different types of antifreeze proteins in various polar and subpolar fishes represents a remarkable example of cold adaptation, but how these unique proteins arose is unknown. We have found that the antifreeze glycoproteins (AFGPs) of the predominant Antarctic fish taxon, the notothenioids, evolved from a pancreatic trypsinogen. We have determined the likely evolutionary process by which this occurred through characterization and analyses of notothenioid AFGP and trypsinogen genes. The primordial AFGP gene apparently arose through recruitment of the 5′ and 3′ ends of an ancestral trypsinogen gene, which provided the secretory signal and the 3′ untranslated region, respectively, plus de novo amplification of a 9-nt Thr-Ala-Ala coding element from the trypsinogen progenitor to create a new protein coding region for the repetitive tripeptide backbone of the antifreeze protein. The small sequence divergence (4–7%) between notothenioid AFGP and trypsinogen genes indicates that the transformation of the proteinase gene into the novel ice-binding protein gene occurred quite recently, about 5–14 million years ago (mya), which is highly consistent with the estimated times of the freezing of the Antarctic Ocean at 10–14 mya, and of the main phyletic divergence of the AFGP-bearing notothenioid families at 7–15 mya. The notothenioid trypsinogen to AFGP conversion is the first clear example of how an old protein gene spawned a new gene for an entirely new protein with a new function. It also represents a rare instance in which protein evolution, organismal adaptation, and environmental conditions can be linked directly.
Keywords: repetitive sequences, gene duplication, environmental selection/de novo amplification
Members of a single teleost suborder, Notothenioidei, overwhelmingly dominate the fish fauna of the freezing (−1.9°C) coastal regions of the Antarctic Ocean in terms of number of species (>50%) (1–3) and biomass (90–95%) (4, 5). Their vast ecological success is linked to ensured survival by the presence of special blood-borne antifreeze glycoproteins (AFGPs; ref. 6). Antifreeze proteins prevent freezing of the body fluids of teleosts, whose equilibrium freezing point (−0.7 to −1°C) is significantly higher than that of seawater (−1.9°C), by adsorbing to small ice crystals in the body and inhibiting their growth (6–9). Besides AFGPs, there are three other structurally different types of antifreeze proteins from various polar and subpolar fishes (10, 11), suggesting that these unique proteins evolved independently at least four times. How these unique proteins evolved in these fishes has remained one of the most important but unanswered questions in the area of antifreeze research. The notothenioid AFGPs exist as a family of at least eight isoforms of different sizes all composed of a simple glycotripeptide repeat, (Thr-Ala/Pro-Ala)n, with the disaccharide galactose-N-acetylgalactosamine attached to each Thr (6, 10), and the dipeptide Ala-Ala at the N terminus. The smallest (n = 4, Mr 2,600) and the largest (n = 55, Mr 34,000) are named AFGP8 and AFGP1, respectively; many other intermediate sizes besides the eight originally described have been subsequently identified (12, 13). Collectively they are maintained at very high circulatory levels of 30–35 mg/ml (6, 10). Notothenioid AFGPs are encoded by large gene families in which each member gene encodes a large polyprotein precursor containing multiple AFGP molecules (12, 14). The first AFGP gene characterized is from the Antarctic notothenioid Notothenia coriiceps, and the AFGP polyprotein precursor it encodes contains 46 AFGP molecules (44 AFGP8 and 2 AFGP7 isoforms) linked in direct tandem by highly conserved three-residue spacers, Leu-Ile/Asn-Phe, which serve as posttranslational cleavage sites to yield the individual AFGP peptides (14). This multigene, multiple AFGP copies per gene organization provides an extremely large gene dosage that undoubtedly contributes to the high circulatory abundance of the protein. The tripeptide repeating structure of the AFGP isoforms and the highly repetitive nature of the AFGP polyprotein gene structure suggest that extensive duplications of an ancestral 9-nt Thr-Ala-Ala coding element had occurred to give rise to AFGPs. However, the origin of such a coding element and the means by which the first AFGP gene was formed are unknown. In searching the GenBank database, we found that the 3′ flanking sequence of the N. coriiceps AFGP gene starting from the termination codon to about 100 nt downstream is about 80% identical to the coding sequence of the C terminus (50 residues) of the trypsinogen cDNA of the Atlantic plaice, suggesting that notothenioid AFGP gene and trypsinogen gene are somehow related. To understand the precise nature of this relationship, AFGP and trypsinogen genes from the giant Antarctic notothenioid, Dissostichus mawsoni were isolated and analyzed. We report here our studies that led to the elucidation of the trypsinogen origin of notothenioid AFGPs and the evolutionary process by which an ancestral trypsinogen gene was transformed to an AFGP gene.
MATERIALS AND METHODS
Characterization of AFGP Genes.
Specimens of the giant Antarctic notothenioid D. mawsoni were caught by winched cable and hook at about 300–500 m in McMurdo Sound, Antarctica. A D. mawsoni genomic library was constructed with liver DNA using the phage vector λFIXII (Stratagene). About a half-million plaque-forming units of the primary library were screened with a 32P-labeled 3.3-kb PstI fragment of the N. coriiceps-AFGP gene (14) by the in situ filter hybridization method (15). Of 125 putative positive clones from the screening, several were randomly selected for analyses. The single AFGP-positive TaqI fragment from each phage clone was subcloned into the plasmid pBluescriptII KS(−) (Stratagene). Nested unidirectional deletions of the subclone insert were generated by exonuclease III digestion (15) and sequenced. Sequence upstream of the 5′ TaqI site was obtained by PCR-amplification of the recombinant phage DNA with the T3 promoter primer in the phage vector and an AFGP-specific primer, cloned into plasmid pCRII (TA Cloning Kit, Invitrogen) and sequenced.
Amplification of AFGP Genes from Genomic DNA.
One microgram of genomic DNA from each of three Antarctic notothenioids, D. mawsoni, N. coriiceps, and Pagothenia borchgrevinki was amplified for AFGP gene sequences by PCR using as primers the 5′ untranslated region (UTR) sequence (cTryp-AF5′), and a 30-nt 3′ primer (cTryp-AF3′) that anneals to a site immediately ahead of the putative polyadenylylation signal site in the 3′ UTR of the AFGP cDNA (see below). A Southern blot (15) of the PCR products was hybridized to an AFGP gene probe that contains only the repetitive Thr-Ala-Ala coding sequence.
Characterization of Trypsinogen and AFGP cDNAs.
Both trypsinogen and AFGP cDNAs were obtained by reverse transcription–PCR (RT-PCR) of pancreatic RNA. Total RNA from the pancreas-associated pyloric caecal mesentery of D. mawsoni was isolated (Ultraspec RNA Isolation System, Biotecx Laboratories, Houston), and poly(A)+ RNA was then isolated from the total RNA (PolyATtract mRNA Isolation System, Promega). About 100 ng of poly(A)+ RNA was reverse-transcribed into first-strand cDNA (Superscript Preamplification System, Life Technologies, Gaithersburg, MD) to which a 5′ anchor oligonucleotide [5′ RACE (Rapid Amplification of cDNA Ends) System, CLONTECH] was ligated. The 5′ portion of trypsinogen cDNA was obtained by 5′ RACE (i.e., PCR-amplification of the first-strand cDNA using the anchor primer and a primer designed from the 3′ flanking sequence of AFGP gene that shares sequence identity with plaice trypsinogen cDNA). The 5′ RACE-PCR products were cloned as a pool into pCRII, and one of the clones was isolated and sequenced. The first 27 nt of the trypsinogen cDNA (5′ UTR) was used to fashion a primer, cTryp-AF5′, and used with a 32-mer oligo(dT) primer to generate the full-length trypsinogen cDNA by RT-PCR amplification of pancreas poly(A)+ RNA, cloned into pCRII and sequenced.
Two partial AFGP cDNAs covering the 5′ and 3′ end were generated. The 5′ cDNA was obtained by RT-PCR primed by the 27-nt cTryp-AF5′ (site found in AFGP gene; see Results) and a 27-nt primer that anneals to AFGP spacer sequence. The 3′ cDNA was obtained by RT-PCR using a 29-nt AFGP-spacer 5′ primer and the 30-nt 3′ primer, cTryp-AF3′. The products were cloned into pCRII and sequenced.
Characterization of Trypsinogen Gene.
The genomic DNA of D. mawsoni was amplified by PCR with the cTryp-AF5′ and the cTry-AF3′ primers, which will amplify both AFGP genes and trypsinogen genes. Trypsinogen PCR products were identified by probing a Southern blot of the PCR products with the trypsinogen cDNA. A trypsinogen-positive product of 2.8 kbp was found, recovered from the gel, cloned into pCRII, and sequenced.
RESULTS
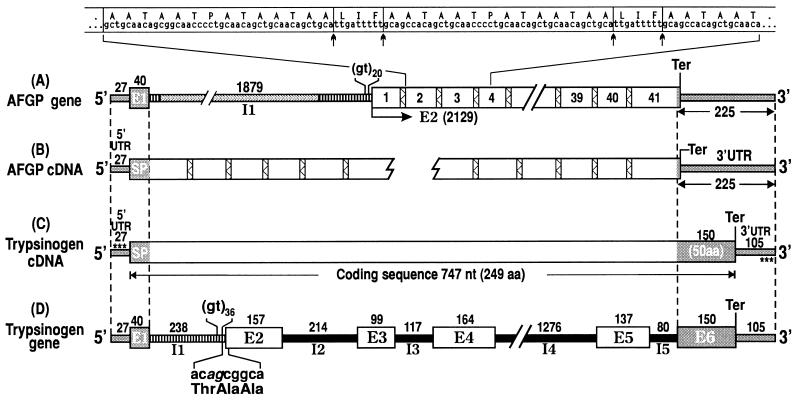
Fig. 1 shows the structures of the characterized AFGP and trypsinogen genes and cDNAs from the giant Antarctic notothenioid D. mawsoni, and their regions of sequence identity.
Figure 1.
Structures of AFGP and trypsinogen genes and cDNAs from Antarctic notothenioid fish D. mawsoni. (A) AFGP gene in genomic clone Dm1A. The AFGP polyprotein coding region (2129 nt) contains 41 copies of AFGP coding sequences (boxed numbers) tandemly linked by highly conserved 9-nt spacers (bars filled with zigzagged lines). Nucleotide and translated amino acid sequence covering copies 2, 3, and (partly) 4 (extended above gene structure) is given. Posttranslational cleavage (↑ ↑) of spacers produces the mature AFGPs. Exon–intron boundaries were determined after two partial AFGP cDNAs were characterized (B). 5′ UTR and signal peptide (SP) sequence are encoded by E1, and AFGP polyprotein by E2. The single, intervening intron I1 is 1879 nt. (C) Trypsinogen cDNA contains a 27-nt 5′ UTR, 747 nt of pretrypsinogen coding sequence, and a 105-nt 3′ UTR. (D) Trypsinogen gene contains six exons (E1-E6) and five introns (I1-I5) with lengths in nt as indicated. Three regions of sequence identities are found on alignment of all four structures. The two pairs of dashed lines delimit the 5′ (67-nt) and 3′ (225-nt) end regions of sequence identities (gray regions) with the segments of the gene or encoded protein each represents, as labeled. The third region of identity is indicated by striped bars; I1 of trypsinogen gene (D) is found in two segments in I1 of AFGP gene (A) and both introns contain repetitive (gt)n at its 3′ end. The 9-nt element in trypsinogen gene that translates into the repeat unit of AFGP peptide backbone, Thr-Ala-Ala, straddles I1 and E2 (splice sequence in italics), as shown in D. Asterisks in C (trypsinogen cDNA) indicate primer sequences cTryp-AF5′ and cTryp-AF3′ used for RT-PCR of AFGP cDNA and trypsinogen gene. Structures are not necessarily drawn to scale.
Notothenioid AFGP Gene Structure.
The complete structure of the AFGP gene from one of the characterized clones, Dm1A (accession no. U58944U58944), and the structure of the 5′ and 3′ AFGP cDNAs (accession nos. U58867U58867, U58868U58868) are depicted in Figs. 1 A and B, respectively. The AFGP polyprotein coding region (2129 nt) in the Dm1A AFGP gene contains 41 copies of AFGP coding sequences that encode four different sized isoforms, linked in tandem by the conserved three-residue spacer, LIF or LNF, similar to that reported for the Nc-AFGP gene from the related N. coriiceps (13). Immediately 5′ to the AFGP coding region is a long stretch of repetitive gt sequences (gt20), which was assigned as part of the presumptive signal peptide in the Nc-AFGP gene (14). However, comparing the AFGP gene and cDNA sequences (Fig. 1 A and B) established that the (gt)n-region in fact comprises the 3′ end of the single intron (I1), which is 1879 nt in length in Dm1A and which intervenes at the C terminus of the true signal peptide encoded in exon 1 (Fig. 1A). Exon 1 encodes the 5′ UTR (27 nt) and most of the signal peptide (40 nt, encodes 13 amino acids). Exon 2 encodes the rest of the signal peptide (six residues by assigning the putative cleavage site right before the AFGP tripeptide repeats begin), the AFGP polyprotein, and the 3′ UTR. The 3′ UTR sequence is 97% identical to that of the Nc-AFGP gene.
Notothenioid Trypsinogen Gene Structure.
The structures of notothenioid trypsinogen cDNA (accession no. U58945U58945) and gene (accession no. U58835U58835) are shown in Figs. 1 C and D, respectively. The intron/exon boundaries were established by comparing the two sequences. The trypsinogen gene contains six exons (E1-E6) and five introns (I1-I5). Exon 1 encodes a 27-nt 5′ UTR and most of the signal peptide (40 nt, 13 amino acids). Exon 2 encodes the remaining two amino acids of the trypsinogen signal peptide (cleavage site inferred from the other trypsinogen sequences in the database). The rest of exon 2 through exon 6 encode a 234-residue trypsinogen and the 3′ UTR.
Alignment of Notothenioid AFGP and Trypsinogen Genes.
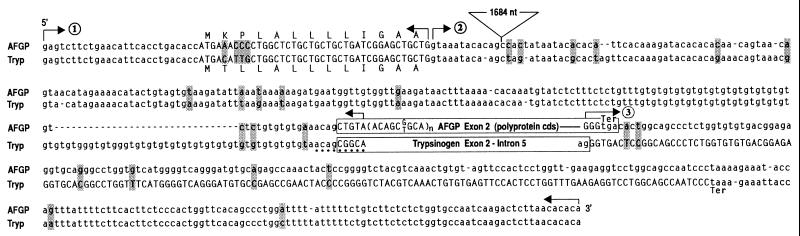
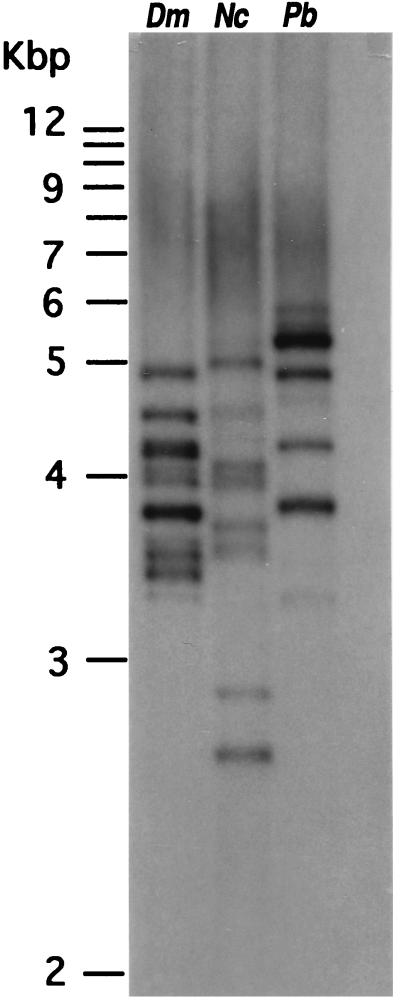
Alignment of the four notothenioid AFGP and trypsinogen gene and cDNA structures showed three regions of sequence identity between the two genes (Figs. 1 A–D). First, exon 1 (5′ UTR and signal peptide) of the AFGP gene and trypsinogen gene are 94% identical in sequence. Thus, the AFGP gene utilizes the 5′UTR and signal peptide coding sequence of the trypsinogen gene. Second, the 3′ end region of AFGP gene starting from the penultimate codon through 3′ UTR (255 nt) is 96% identical to the last exon (E6) of trypsinogen gene. Thus, the AFGP gene utilizes trypsinogen E6 as 3′ UTR. Third, the entire trypsinogen intron 1 (238 nt) is present as two segments within the single AFGP intron with an overall sequence identity of 93%. The first 12 nt of trypsinogen I1 inclusive of 5′ splice site correspond to the same in AFGP I1, and the remaining 226 nt inclusive of 3′ splice site and a (gt)36 sequence right before the 3′ splice site correspond to the (gt)20-bearing 3′ end of AFGP I1. Thus, AFGP gene intron 1 is the same as trypsinogen gene I1 plus a large (1.7-kbp) insertion. Immediately 3′ to the (gt)36 sequence in the trypsinogen gene and straddling the splice junction of I1 and E2, a 9-nt element, acagcggca (splice sequence in italics) is found that translates into Thr-Ala-Ala (Fig. 1D and Fig. 2), the building block of the repetitive tripeptide sequence of AFGP. The nucleotide sequence of the three regions of sequence identity between the two genes are shown in Fig. 2. The trypsinogen/AFGP hybrid gene structure is found to be present in the extant AFGP genes of the notothenioids as shown by the multiple AFGP-positive bands on a Southern blot of PCR-amplification products from the genomic DNA of three different notothenioids using the common trypsinogen/AFGP 5′ UTR (cTryp-AF5′) and 3′ UTR (cTryp-AF3′) primers (Fig. 3) and hybridized to an AFGP-specific probe that contains only (Thr-Ala-Ala)n coding sequence.
Figure 2.
Alignment of D. mawsoni AFGP and trypsinogen (Tryp) genomic sequences showing the three regions of high sequence identity between the two genes. ①, 5′ UTR (lowercase) and signal peptide coding sequences (uppercase; translated amino acids also shown) are 94% identical; ②, intron I sequences (lowercase) are 93% identical (position of the extra 1684-nt AFGP intron sequence is indicated); and ③, AFGP penultimate codon plus 3′UTR sequence (lowercase) is 96% identical to trypsinogen exon 6 (uppercase) and 3′ UTR (lowercase). Positions of gene-specific sequences are boxed. The small number of nucleotide differences in the three regions are highlighted. The dots underscore the 9-nt Thr-Ala-Ala coding element (acagcggca) in trypsinogen that might have been amplified to give rise to the repetitive tripeptide coding sequence of AFGP (in parentheses).
Figure 3.
Southern blot of products from PCR-amplification of genomic DNA from three different notothenioids, using common 5′ and 3′ trypsinogen/AFGP primers (sites indicated by asterisks in Fig. 1C) and hybridized to an AFGP-specific probe containing the repetitive AFGP coding sequence only. The multiple AFGP-positive bands indicate that members of notothenioid AFGP gene families all have the hybrid AFGP/trypsinogen gene structure. Dm, Dissostichus mawsoni; Nc, Notothenia coriiceps; Pb, Pagothenia borchgrevinki.
DISCUSSION
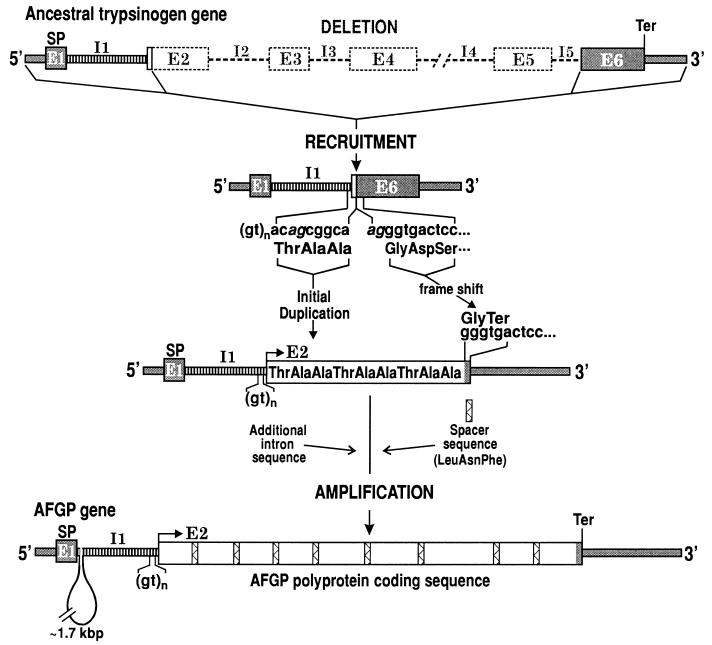
Given the common sequence elements between notothenioid trypsinogen and AFGP genes, and the occurrence of a Thr-Ala-Ala coding element in the trypsinogen gene, the primordial AFGP gene could have risen from an ancestral trypsinogen gene by a combined process of partial gene recruitment and de novo amplification depicted in Fig. 4. E1 (5′ UTR and signal peptide), I1, and several nucleotides of trypsinogen E2 inclusive of the 9-nt Thr-Ala-Ala coding element were recruited to form the 5′ portion of the AFGP gene. A deletion removed the rest of trypsinogen E2 through I5, linking E6 inclusive of the 3′ UTR to the 9-nt Thr-Ala-Ala coding element, and de novo amplification of the coding element gave rise to an entirely new coding region that encodes the repetitive tripeptide backbone of AFGP. The deletion, recruitment, and amplification events did not need to occur in the order given. Indeed, an AFGP/trypsinogen hybrid protein coding region formed by some amount of duplication of the 9-nt Thr-Ala-Ala coding element before bulk deletion of trypsinogen sequence might in fact be a more stable structure for the evolving gene than large deletion first and amplification later. In any case, these DNA rearrangement and amplification events together led to a frameshift resulting in a termination codon (tga) at the start of the recruited trypsinogen E6, and converting it into the penultimate codon that encodes the last amino acid, Gly (ggg; 1 g from splice sequence, 2 from E6) and the 3′ flanking region (or 3′ UTR) of the new gene (Fig. 4). The same penultimate codon (ggg) is found in all notothenioid AFGP genes sequenced (refs. 13 and 14; Fig. 2).
Figure 4.
Likely mechanism by which an ancestral trypsinogen gene was transformed into an AFGP gene. The 5′ end (E1, I1, and small segment of E2) and the 3′ end (I5 3′ splice site and E6) of trypsinogen gene were recruited and linked, and the remainder of the gene deleted (dashed lines and boxes). The Thr-Ala-Ala coding element was duplicated, presumably via slippage at the repetitive (gt)n sequence during replication. The recruited E1 provided the 5′ UTR and signal peptide sequences for the new AFGP gene. The deletion, linking, and amplification events led to a 1-nt frameshift resulting in a termination codon (tga) at the start of the recruited trypsinogen E6 and converting it into the 3′ flanking sequence of the AFGP gene. The spacer sequence (bars filled with zigzagged lines) and additional I1 sequence might be existing sequence in the trypsinogen progenitor gene or acquired through recombinatory events. The Thr-Ala-Ala coding duplicants plus a spacer became amplified de novo to form the new AFGP polyprotein coding region. The regions of identity are illustrated as in Fig. 1. Splice sites in trypsinogen gene are given in italics.
The initial duplication of the 9-nt Thr-Ala-Ala coding element could be a result of slippage replication (16) at the repetitive (gt)n sequence immediately upstream during DNA replications, causing the Thr-Ala-Ala coding element to be copied more than once. Subsequent amplifications could occur through slippage replication or unequal crossing over (17, 18) of the new duplicants. Evidence for amplification of the trypsinogen Thr-Ala-Ala coding element to give rise to the repetitive AFGP polyprotein coding sequence is provided by a striking correspondence between its nucleotide sequence, aca(Thr)-gcg(Ala)-gca(Ala), and those of the tripeptide repeats in extant AFGP genes. In the Dm1A AFGP gene, the preponderance of aca in coding Thr (137/170, 81%), and gca in coding the second Ala (162/171, 95%) strongly indicates that they are duplicants or descendants of the ancestral trypsinogen codons. The first Ala in the tripeptide is coded mostly by gct (65/107, 61%) and the remainder by gcg (38/107, 36%). It is likely that gct, rather than gcg, was the codon for the first Ala in the ancestral trypsinogen Thr-Ala-Ala coding element. Nucleotide substitutions at this codon led to either no change in the amino acid (gct to gcg, Ala), or a Pro for Ala replacement (gct to cct) observed at this Ala position in the antifreeze protein. The occurrence of the latter (gct to cct) is supported by the almost exclusive use of cct in coding the Pro (63/64, 96%) in the Thr-Pro-Ala repeats in the Dm1A gene, as well as in other characterized notothenioid AFGP genes (13, 14).
The origin of the three-residue spacer sequence (Leu/Phe-Ile/Asn-Phe) is unclear at this point. It could be a preexisting element that adjoined the Thr-Ala-Ala coding element in the recruited ancestral trypsinogen gene, or was acquired through recombinatory events early in the duplications of the Thr-Ala-Ala coding element. The two parts then could have subsequently amplified together to give rise to the extant polyprotein gene structure (Fig. 4). If amplification of the 9-nt Thr-Ala-Ala coding element occurred before deletion of trypsinogen sequence, the acquisition of the tripeptide spacer would provide the cleavage site to excise the first AFGP molecule from the AFGP/trypsinogen hybrid protein before the stop codon (tga) was appropriately established for the evolving AFGP gene. Like the spacer sequence, preexisting sequence or recombinatory events could account for the additional sequence (≈1.7 kbp) in the intron of the AFGP gene. The trypsinogen progenitor was lost once it was converted into the first AFGP gene, and the apparent absence of spacer-like sequence or extra intron 1 sequence in extant trypsinogen genes thus does not preclude the possibility that they might be present in the trypsinogen progenitor before its transformation into an AFGP gene. To summarize, through recruitment and linking of the 5′ and 3′ portions of a trypsinogen gene that supplied the secretory signal and the 3′ flanking region (or 3′ UTR), respectively, and de novo expansion of a 9-nt Thr-Ala-Ala coding element in the middle of the new structure to form the AFGP coding region, the first functional AFGP gene was formed. The hybrid trypsinogen/AFGP gene structure, like that of Dm1A, is confirmed to be present in current members of AFGP gene families across Antarctic notothenioid species (Fig. 3).
Deciphering the evolutionary process of notothenioid AFGP gene from a trypsinogen gene was made possible by the high degree of nucleotide identities (93–96%) in both the coding and noncoding sequences between the two genes, as well as the close correspondence between the candidate 9-nt ancestral Thr-Ala-Ala coding element in trypsinogen gene and the repetitive AFGP tripeptide coding sequences, all of which indicate that the trypsinogen to AFGP conversion was a recent event. The small divergence (7%) between AFGP and trypsinogen intron 1 sequences particularly supports the recent evolution of the notothenioid AFGP as intron sequences are under no constraint to remain conserved. There are no definitive nuclear gene sequence divergence rates for teleosts available in the literature to estimate the time of divergence between notothenioid trypsinogen and AFGP genes. Using teleost (salmon) mitochondrial DNA divergence rates, 0.5–0.9% per million years (myr; ref. 19), the 4–7% sequence divergence between the two notothenioid genes translates into a divergence time of about 5–14 myr. Evolution of AFGPs at 5–14 million years ago (mya) is remarkably consistent with the estimated mid-Miocene (10–14 mya) time frame during which the Antarctic Ocean cooled to freezing based on paleotemperatures inferred from oxygen isotopic ratios of sea bottom planktonic deposits (20–22). It is also highly consistent with the molecular phylogeny of notothenioids based on mitochondrial 12S and 16S ribosomal RNA gene sequences, which places the main phyletic divergence of the five AFGP-bearing notothenioid families at about 7–15 mya (23). These time frames are not entirely definitive, since paleoceanographic temperatures derived from oxygen isotopic methods were subject to different interpretations, and hypotheses of phyletic relationships among notothenioids differ depending on whether morphological or molecular data are subject to cladistic analysis (3, 23). However, since the driving force for AFGP evolution came from the onset of freezing conditions in the Antarctic waters, and emergence of AFGPs undoubtedly enabled the phyletic radiations of the benthic ancestral notothenioids stock (3) into ice-laden pelagic and surface habitats where they reside today, it is reasonable to expect that the chronology of these three events would overlap. The remarkable agreement in the estimated Miocene times of AFGP gene evolution in this study, the freezing of the Antarctic Ocean inferred from paleoceanographic studies (20–22), and the main phyletic divergence of notothenioids based on molecular phylogeny (23) argue strongly that this time frame could not be due to mere coincidence, but is, in fact, reliable.
A pertinent question is why a pancreatic enzyme protein gene was selected for conversion to the new ice-binding protein. The intestinal fluids of Antarctic notothenioids are known to contain high concentration of AFGPs (24) which serve to inhibit the growth of ice crystals that inevitably enter through food and seawater ingestion (9, 24). Conversion of an existing pancreatic enzyme gene into AFGP gene and expressing it in the pancreas is both positionally and temporally logical as AFGPs thus could reach the digestive tract, simultaneously with pancreatic enzymes, to prevent the intestinal fluid from freezing while the enzymes perform digestive functions. AFGPs continue to be expressed in the notothenioid pancreas today as verified by the production of AFGP cDNAs by RT-PCR amplification of pancreatic mRNA in this study, and the expression appears to be at high levels as indicated by the high intensity of hybridization to an AFGP-specific probe on a Northern blot of pancreatic mRNA (data not shown). It is possible that the notothenioid AFGP gene evolved and became expressed in the pancreas first to protect the intestinal fluid, as it was readily susceptible to freezing through daily intake of ice-associated food or seawater, and later, the expression extended to the liver, which became the synthetic site to provide circulatory antifreeze (25) for freezing avoidance in other extracellular fluid compartments.
The elucidation of how a notothenioid pancreatic trypsinogen gene was transformed into an AFGP gene provides the first clear and plausible evolutionary process by which one of the four known types of antifreeze proteins arose. The antifreeze peptides (type II AFPs) from the sea raven, herring, and smelt share partial protein sequence identity with the carbohydrate recognition domain of C-type lectins or similar domains in lectin-like proteins from other organisms, suggesting evolutionary relatedness (26). Lectins in type II AFP-bearing fishes have not been characterized, and if they indeed gave rise to the type II AFPs in these fishes, it would represent an example of gene recruitment and expression of the same or a very similar protein to perform a different function, much like the recruitment of cellular enzyme genes such as lactate dehydrogenase and others, and express them at high levels to form the lens crystallins (27–30). Evolution of notothenioid AFGP genes represents another evolutionary innovation—recruitment of segments of an existing protein gene plus de novo amplification of a short DNA sequence to spawn a novel protein with a new function. Despite the apparently recent notothenioid AFGP gene evolution, powerful environmental selectional pressure—that is, the threat of freezing death once the Antarctic water reached perennial freezing temperatures—may have driven rapid intragene and whole gene duplications. These processes could readily occur for simple repetitive sequences like the AFGPs, leading to the large AFGP polyprotein gene families we see today.
Acknowledgments
This work is supported in part by National Science Foundation Grant OPP-93–17629 to A.L.D.
ABBREVIATIONS
- AFGP
antifreeze glycoprotein
- myr
million years
- mya
million years ago
- RT-PCR
reverse transcription–PCR
- UTR
untranslated region
Footnotes
References
- 1.Gon O, Heemstra P C. Fishes of the Southern Ocean. Grahamstown, South Africa: JLB Smith Institute of Ichthyology; 1990. [Google Scholar]
- 2.Hubold G. In: Biology of Antarctic Fish. di Prisco G, Maresca M, Tota B, editors. Berlin: Springer; 1991. pp. 3–22. [Google Scholar]
- 3.Eastman J T. Antarctic Fish Biology: Evolution in a Unique Environment. San Diego: Academic; 1993. [Google Scholar]
- 4.DeWitt H H. In: Antarctic Map Folio Series, Folio 15. Bushnell V C, editor. New York: Am. Geogr. Soc.; 1971. pp. 1–10. [Google Scholar]
- 5.Ekau W. Antarct Sci. 1990;2:129–137. [Google Scholar]
- 6.DeVries A L. Comp Biochem Physiol B. 1988;90:611–621. [Google Scholar]
- 7.Raymond J A, DeVries A L. Proc Natl Acad Sci USA. 1977;74:2589–2593. doi: 10.1073/pnas.74.6.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.DeVries A L. Philos Trans R Soc London B. 1984;304:575–588. [Google Scholar]
- 9.DeVries A L, Cheng C-H C. In: Water and Life. Somero G N, Osmond C B, Bolis C L, editors. Berlin: Springer; 1992. pp. 303–315. [Google Scholar]
- 10.DeVries A L. Comp Biochem Physiol A. 1982;73:627–640. [Google Scholar]
- 11.Davies P L, Hew C L. FASEB J. 1990;4:2460–2468. doi: 10.1096/fasebj.4.8.2185972. [DOI] [PubMed] [Google Scholar]
- 12.Cheng C-H C. In: Gene Expression and Manipulation in Aquatic Organisms. Ennion S, Goldspink G, editors. Cambridge, UK: Cambridge Univ. Press; 1996. pp. 1–20. [Google Scholar]
- 13.Chen L, DeVries A L, Cheng C-H C. Proc Natl Acad Sci USA. 1997;94:3817–3822. doi: 10.1073/pnas.94.8.3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hsiao K C, Cheng C-H C, Fernandes I E, Detrich H W, DeVries A L. Proc Natl Acad Sci USA. 1990;87:9265–9269. doi: 10.1073/pnas.87.23.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 16.Dover G A, Tautz D. Philos Trans R Soc London B. 1986;312:275–289. doi: 10.1098/rstb.1986.0007. [DOI] [PubMed] [Google Scholar]
- 17.Maeda N, Smithies O. Annu Rev Genet. 1986;20:81–108. doi: 10.1146/annurev.ge.20.120186.000501. [DOI] [PubMed] [Google Scholar]
- 18.Lewin B. Gene IV. New York and Cell Press, Cambridge, MA: Oxford Univ. Press; 1990. pp. 497–517. [Google Scholar]
- 19.Martin A P, Palumbi S R. Proc Natl Acad Sci USA. 1993;90:4087–4091. doi: 10.1073/pnas.90.9.4087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kennett J P. J Geophys Res. 1977;82:3843–3860. [Google Scholar]
- 21.Kennett J P. Marine Geology. Englewood, NJ: Prentice–Hall; 1982. [Google Scholar]
- 22.Clarke A. In: Antarctic Ecosystems: Ecological Change and Conservation. Kerry K R, Hempel G, editors. Berlin: Springer; 1990. pp. 9–22. [Google Scholar]
- 23.Bargelloni L, Ritchie P A, Patarnello T, Battaglia B, Lambert D M, Meyer A. Mol Biol Evol. 1994;11:854–863. doi: 10.1093/oxfordjournals.molbev.a040168. [DOI] [PubMed] [Google Scholar]
- 24.O’Grady S M, Ellory J C, DeVries A L. J Exp Biol. 1983;104:149–162. [Google Scholar]
- 25.Hudson A P, DeVries A L, Haschemeyer A V E. Comp Biochem Physiol B. 1979;62:179–183. [Google Scholar]
- 26.Ewart K V, Fletcher G L. Mol Mar Biol Biotechnol. 1993;2:20–27. [PubMed] [Google Scholar]
- 27.Wistow G, Piatigorsky J. Science. 1987;236:1554–1556. doi: 10.1126/science.3589669. [DOI] [PubMed] [Google Scholar]
- 28.Wistow G, Anderson A, Piatigorsky J. Proc Natl Acad Sci USA. 1990;87:6277–6280. doi: 10.1073/pnas.87.16.6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piatigorsky J, Wistow G. Science. 1991;252:1078–1079. doi: 10.1126/science.252.5009.1078. [DOI] [PubMed] [Google Scholar]
- 30.Zinovieva R D, Tomarev S I, Piatigorsky J. J Biol Chem. 1993;268:11449–11455. [PubMed] [Google Scholar]