Abstract
Antarctic notothenioid fishes and several northern cods are phylogenetically distant (in different orders and superorders), yet produce near-identical antifreeze glycoproteins (AFGPs) to survive in their respective freezing environments. AFGPs in both fishes are made as a family of discretely sized polymers composed of a simple glycotripeptide monomeric repeat. Characterizations of the AFGP genes from notothenioids and the Arctic cod show that their AFGPs are both encoded by a family of polyprotein genes, with each gene encoding multiple AFGP molecules linked in tandem by small cleavable spacers. Despite these apparent similarities, detailed analyses of the AFGP gene sequences and substructures provide strong evidence that AFGPs in these two polar fishes in fact evolved independently. First, although Antarctic notothenioid AFGP genes have been shown to originate from a pancreatic trypsinogen, Arctic cod AFGP genes share no sequence identity with the trypsinogen gene, indicating trypsinogen is not the progenitor. Second, the AFGP genes of the two fish have different intron–exon organizations and different spacer sequences and, thus, different processing of the polyprotein precursors, consistent with separate genomic origins. Third, the repetitive AFGP tripeptide (Thr-Ala/Pro-Ala) coding sequences are drastically different in the two groups of genes, suggesting that they arose from duplications of two distinct, short ancestral sequences with a different permutation of three codons for the same tripeptide. The molecular evidence for separate ancestry is supported by morphological, paleontological, and paleoclimatic evidence, which collectively indicate that these two polar fishes evolved their respective AFGPs separately and thus arrived at the same AFGPs through convergent evolution.
Keywords: repetitive sequence, gene duplication, sequence convergence, trypsinogen
Various polar and north-temperate marine teleosts had evolved a total of four structurally different types of antifreeze protein, which enabled them to survive and successfully colonize their respective icy, freezing (−1.9°C) habitats (1–4). Surprisingly, phyletically related fishes do not necessarily possess the same type of antifreeze protein (5, 6) or vice versa for unrelated fishes (7). The most striking example of evolution of the same type of antifreeze protein in unrelated fishes is the near-identical antifreeze glycoproteins (AFGPs) in the Antarctic notothenioid fishes and several northern cods of the family Gadidae (8, 9). Antarctic notothenioids are modern perciforms in the superorder Acanthopterygii, while northern cods are gadiforms in the less phyletically derived superorder Paracanthopterygii (10). AFGPs in both fishes comprise a family of at least eight size/compositional isoforms all composed of a simple glycotripeptide repeat, (Thr-Ala/Pro-Ala)n, with each Thr O-linked to the disaccharide, galactosyl-N-acetylgalactosamine, and n = 4 to ≥ 55 (1–4, 8, 9). AFGPs of cods differ only in the presence of an occasional Arg-for-Thr substitution (8, 11).
Notothenioid AFGPs are encoded by a large gene family, and each member gene encodes a large polyprotein precursor containing many AFGP molecules (as many as 46) linked in tandem by highly conserved tripeptide spacers (Leu/Phe-Ile/Asn-Phe), which are cleaved posttranslationally to produce the mature AFGPs (12, 13). Notothenioid AFGP genes have recently been discovered to evolve from a trypsinogen gene through recruitment of segments of the trypsinogen sequence, plus de novo amplification of a 9-nt Thr-Ala-Ala coding element from the trypsinogen progenitor, to form a new coding region for the repetitive tripeptide backbone of AFGPs (12). No studies of the AFGP genes of northern cods have been reported to date. To determine how northern cod AFGPs are encoded and how two unrelated fish taxa arrive at essentially the same AFGP molecules, we have characterized the AFGPs and AFGP genes from the Arctic cod Boreogadus saida (Svalbard, Norway) and compared them to those of the Antarctic notothenioid Dissostichus mawsoni (McMurdo Sound, Antarctica).
MATERIALS AND METHODS
Purification and Analysis of AFGPs.
AFGPs were purified from B. saida and D. mawsoni plasma by DEAE-cellulose ion exchange chromatography (8). Purified AFGPs were fluorescently labeled and electrophoresed on a 15–20% nondenaturing, gradient polyacrylamide gel (8) for size heterogeneity comparison. Amino acid compositions of AFGPs were analyzed at the Biotechnology Center (University of Illinois).
D. mawsoni AFGP Gene Characterization.
Genomic library construction and screening, and subcloning and sequencing of AFGP gene sequences were carried out as reported (12).
B. saida AFGP Gene Characterization.
A partial library enriched for AFGP sequences was constructed for B. saida. Spleen DNA was completely digested with MboI, and a cluster of AFGP-positive (to Nc-AFGP probe, a 3.3-kbp PstI AFGP gene fragment from Notothenia coriiceps; ref. 13) fragments between 1 and 4 kbp were recovered from 0.7% low-melting agarose gel (see Results) and cloned into the XhoI site of the insertion phage vector λZAPExpress (Stratagene). Phagemid (pBK-CMV) subclones were excised from the recombinant phages with the ExAssist helper phage (Stratagene) and screened by colony hybridization (14) with the Nc-AFGP probe. Nested sets of unidirectional deletions of the inserts of three positive subclones were generated using exonuclease III (14) and sequenced.
Northern Blot Analysis of B. saida AFGP mRNA.
B. saida liver total RNA was extracted with Ultraspec RNA Isolation Reagent (Biotecx Laboratories, Houston). Poly(A)+ RNA was isolated from total RNA with the PolyATtract mRNA Isolation System (Promega), resolved on 1.2% agarose/2 M formaldehyde gel, vacuum-blotted onto nylon membrane, and hybridized at 60°C to a B. saida AFGP gene probe (from clone Bs3–1; see Results).
B. saida cDNA Characterization.
The 5′ portion of a B. saida AFGP cDNA was obtained by 5′ RACE (Rapid Amplification of cDNA Ends System, CLONTECH) of liver poly(A)+ RNA, using the 5′ anchor primer (provided by the system) and an AFGP-specific primer for amplification. The amplified cDNA was cloned into the plasmid pCRII (TA Cloning Kit, Invitrogen) and sequenced.
Southern Blot Analyses of Genomic DNA.
B. saida and D. mawsoni genomic DNA was completely digested with MboI, resolved on 1% agarose gel, vacuum-blotted onto nylon membrane, and hybridized at 60°C to the Bs3–1 and D. mawsoni AFGP gene probe, respectively.
RESULTS
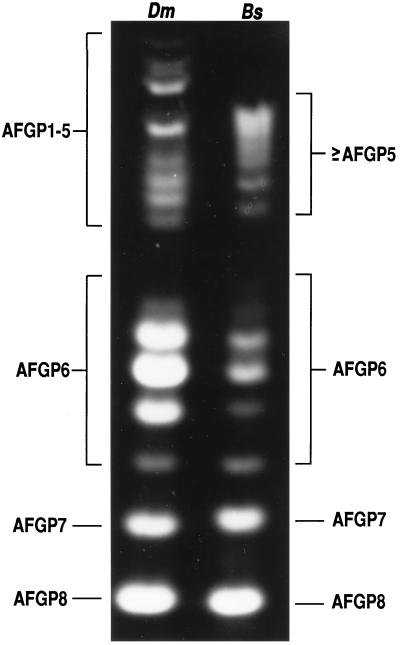
Fig. 1 and Table 1 illustrate the similarities at the protein level between the AFGPs of the Antarctic notothenioid D. mawsoni and the Arctic cod B. saida. The AFGP size isoforms from the two fishes are shown in Fig. 1. Size heterogeneity for the smaller AFGPs 6–8 is identical between the two fishes as indicated by the same number of bands and their identical gel mobilities. The larger sizes, AFGP5 and above, show more variability; the largest AFGP of B. saida is smaller than that of D. mawsoni, corresponding to the size of an AFGP3 in the latter (Fig. 1). AFGP6, which was originally detected as a single size in the notothenioids (1, 2), in fact comprises at least six sizes of isoforms on enhanced staining and gel resolution methods. Similarly, there are more than five isoforms in the large AFGP1–5 size group (Fig. 1). Amino acid analyses of B. saida AFGPs show the presence of the three residues of the tripeptide repeat—Ala, Pro, and Thr—plus a small amount of Arg in all sizes (Table 1). The large AFGP1–5 has the highest Arg content of the three size groups, plus a small amount of Val not found in the others. The stoichiometry of non-Thr (Ala, Pro) to Thr residues for all the sizes is about 2:1 after accounting for the AFGP N terminus dipeptide, Ala/Pro-Ala.
Figure 1.
Polyacrylamide gel of fluorescently labeled AFGPs from Antarctic notothenioid D. mawsoni (Dm) and Arctic cod B. saida (Bs) and the amino acid compositions of the three size groups of Arctic cod AFGPs. The two polar fishes show comparable size heterogeneity, especially in the range of AFGPs 6–8.
Table 1.
Amino acid composition (mol %) of the three AFGP size groups from B. saida
| Size group | Ala | Pro | Thr | Arg | Val |
|---|---|---|---|---|---|
| ≥AFGP5 | 67.0 | 1.2 | 29.8 | 1.2 | 0.8 |
| AFGP6 | 61.0 | 12.3 | 26.1 | 0.6 | — |
| AFGP7,8 | 58.6 | 14.1 | 26.6 | 0.7 | — |
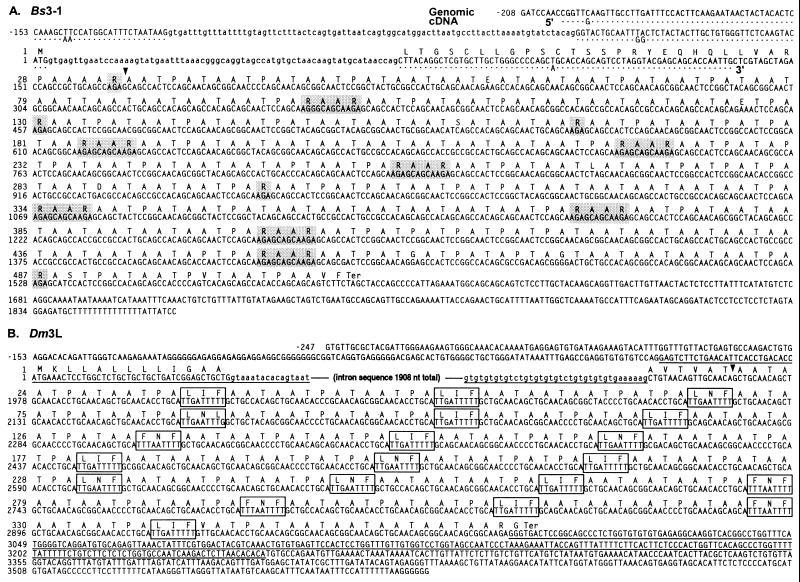
The contiguous sequence (2070 nt) of one of the Arctic cod AFGP genes characterized (subclone Bs3–1) is shown in Fig. 2A. A partial AFGP cDNA (208 nt) corresponding to the 5′ portion of the gene was obtained by 5′ RACE and aligned with the genomic sequence to delineate the exon–intron structure of that region (Fig. 2A). The gene has three exons and two small introns shown schematically in Fig. 3. Intron 1 intervenes in the 5′ flanking sequence (82 nt, −127 to −45), and intron 2 intervenes in the signal peptide coding region (69 nt, +4 to +72). Exon 1 encodes part of the 5′ untranslated region (UTR) (81 nt, −208 to −128), exon 2 encodes the remaining 5′ UTR and the translation start (47 nt, −44 to +3), and exon 3 encodes the signal peptide and the AFGPs (1518 nt, +73 to +1590). The signal peptide is 33 residues in length by assigning the putative cleavage site at residue 33 (Arg, nucleotides +166 to +168). The uninterrupted AFGP coding region (+169 to +1590) that follows encodes a large AFGP precursor containing 156 consecutive tripeptide repeats; 136 are T-A/P-A (Thr-Ala/Pro-Ala) and 20 are R-A-A (Arg-Ala-Ala), plus 3 additional C terminus residues, Ala-Val-Phe. The occurrence of R-A-A leads to the periodic placement of an R or R-A-A-R (Fig. 2A). The mature AFGPs have much less Arg (0.6–1.2%; Fig. 1) than in the AFGP gene sequence (7.5%), suggesting that some of the Arg residues act as cleavable spacers. Thus, the AFGP precursor is a polyprotein, and if all the Arg are cleaved, 13 mature AFGP molecules will be produced, ranging in size from AFGP7 (5 tripeptide repeats) to 1 molecule in the size range of AFGP5 (20 repeats).
Figure 2.
Nucleotide sequence of an AFGP polyprotein gene and the encoded amino acids from B. saida subclone Bs3–1 aligned with a 5′ AFGP cDNA (A), and D. mawsoni subclone Dm3L (B). Dots represent identical nucleotides between the B. saida AFGP gene and cDNA sequences. The introns are shown in lowercase letters. The distinct spacer sequences linking multiple AFGP sequences are indicated in shaded boxes (Bs3–1) or open boxes (Dm3L). ▾, Putative cleavage site for the signal peptide in both genes. The underlined sequences in Dm3L (B) are regions that share high sequence identities with notothenioid trypsinogen gene (12); not all of the similar intron sequence is shown.
Figure 3.
Exon–intron organizations of B. saida and D. mawsoni AFGP genes, showing also the dissimilar signal peptide sequences of the two AFGP polyproteins. The coding region for the signal peptide in each gene is indicated by the shaded areas. ▾, Putative cleavage site for the signal peptide.
For comparison, Fig. 2B shows the sequence (3834 nt) of an AFGP gene (subclone Dm3L) from the Antarctic notothenioid, D. mawsoni. The gene has two exons and a single large intron shown schematically in Fig. 3. The intron (1908 nt) intervenes in the signal peptide coding region (+41 to +1948), and carries a stretch of repetitive (gt)n sequence at its 3′ end, characteristic of notothenioid AFGP genes (12, 13). Exon 1 (67 nt, +1 to +67) encodes a 27-nt 5′ UTR and most of the signal peptide (13 residues), and exon 2 encodes the remainder of the signal peptide (6 residues; cleavage site tentatively assigned at the Thr encoded by +1963 to +1965) and the AFGP polyprotein. The encoded AFGP polyprotein (+1966 to +2994) contains 21 molecules of AFGPs ranging from sizes smaller than AFGP8 (two to three tripeptide repeats) to AFGP6 (six repeats), linked in tandem by the conserved three-residue spacers, L/F-I/N-F/L (Leu/Phe-Ile/Asn-Phe/Leu). The three regions of sequence identity (93–96%) to D. mawsoni trypsinogen gene (12) are underlined (Fig. 2B).
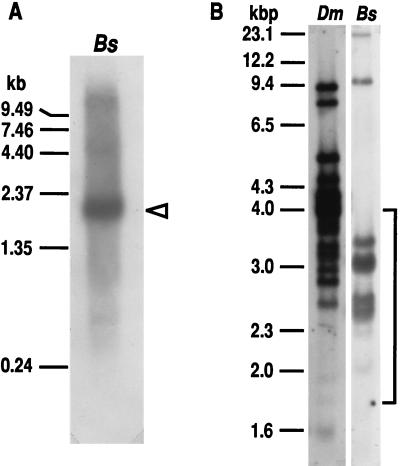
Northern blot analysis of Arctic cod liver poly(A)+ RNA revealed multiple AFGP messages in the size range of 0.5–7 kb (Fig. 4A). The most abundant size, 1.8 kb, approximates the estimated size of the mRNA for Bs3–1 AFGP gene (1.5 kb plus UTRs and the polyA tail), which is consistent with the gene being transcribed and translated as a polyprotein. The intense 1.8-kb band in fact may comprise the messages of similarly sized AFGP genes contained in the cluster of positive MboI restriction fragments between 2.5 and 3.5 kbp in the genomic Southern blot (Fig. 4B); MboI does not cleave within the AFGP coding sequences of Bs3–1 and other Arctic cod AFGP genes (unpublished data) or notothenioid AFGP genes (12, 13), and thus the positive bands contain complete genes. The multiple positive bands in the genome of Arctic cod and Antarctic notothenioid fish indicate the presence of an AFGP multigene family, with that of the notothenioid being much larger (Fig. 4B).
Figure 4.
(A) Northern blot of liver poly(A)+ RNA from Arctic cod (Bs) showing multiple AFGP messages. ◃, Predominant 1.8-kb mRNA. (B) Southern blot of genomic DNA digested with MboI from D. mawsoni (Dm) and B. saida (Bs) showing AFGP multigene family. Bracket indicates the range of DNA fragments recovered for construction of B. saida partial genomic library.
DISCUSSION
Antarctic notothenioid fish and northern cods are phyletically distant (10) but synthesize near-identical AFGPs (refs. 1–4, 8, 9, and 11; Fig. 1). For the Arctic cod, B. saida, the stoichiometry of Ala/Pro to Thr residues of about 2:1 for all its AFGP size isoforms (Table 1) is consistent with the presence of monomeric tripeptide repeats in the protein backbone. The percentage of Pro is very low in the large AFGPs (≥AFGP5), but 10-fold higher in the small AFGPs 6–8 (Fig. 1). This is similar to notothenioid AFGPs, in that Pro-for-Ala substitutions occur in the small AFGPs 6–8, and the large AFGPs 1–5 consist of predominantly Thr-Ala-Ala repeats (1, 2, 15). The only obvious difference between the two groups of AFGPs is that a small amount of Arg is present in Arctic cod AFGPs, as it is for the AFGPs of other northern cods (8, 11). In Alaskan saffron cod (8) and Atlantic tomcod (11), Arg is found to substitute Thr. The carbohydrate moiety in all cases was found to be the same disaccharide, galactosyl-N-acetylgalactosamine (1, 8, 9, 11, 15). In terms of size heterogeneity, D. mawsoni and B. saida have identical AFGPs 6–8 (Fig. 1). Variation is seen in the larger sizes (≥AFGP 5) only (Fig. 1), which is not unexpected as it is also seen between different genera (16, 17) and among individuals of the same species (17) of notothenioids. Thus, except for the presence of small amounts of Arg, the primary structures of AFGPs of notothenioids and B. saida and other northern cods are indistinguishable.
AFGP genes of Arctic cod (Fig. 2A) and notothenioids (refs. 12 and 13; Fig. 2B) both have a polyprotein structure in the AFGP coding region and encode many copies of AFGP linked in tandem by small cleavable spacers. For both fish taxa, the large sizes of the AFGP messages (ref. 13; Fig. 4A), sequences of AFGP cDNAs (ref. 12; unpublished data), and the absence (notothenioid) or reduced number (Arctic cod) of spacer residues in the mature AFGPs indicate that their respective AFGP genes are transcribed and translated into large polyproteins that are posttranslationally cleaved. The near-identical AFGP protein structures from the two fishes have led to suggestions of a common ancestor (18), and the apparent similarities in their AFGP genes appear to support such an argument. However, detailed analyses of the gene substructures and nucleotide sequences, discussed below, provide strong evidence that a common ancestry for these two AFGP gene families is in fact very unlikely.
Notothenioid AFGP genes have been shown to have evolved from a pancreatic trypsinogen gene (12) and, in fact, inherited trypsinogen exon 1 (5′ UTR and signal peptide coding sequence) and intron 1, as well as utilized trypsinogen exon 6 (coding sequence of the 50 C terminus residues plus 3′ UTR) as 3′ UTR. The sequence identity in these regions is very high, 93–96% in all characterized notothenioid AFGP genes (12). By contrast, nucleotide sequences of these regions in the Arctic cod AFGP gene are entirely different from those of the notothenioid AFGP gene. The signal peptide of Arctic cod AFGP polyprotein is entirely different and substantially longer in protein sequence from that of notothenioid AFGP polyprotein (33 vs. 19 residues; Fig. 3). Also, Arctic AFGP gene sequence shares no similarity at all with the trypsinogen cDNA sequence of the closely related Atlantic cod (same family), Gadus morhua (19). Thus, Arctic cod AFGP gene is unrelated to notothenioid AFGP gene or trypsinogen gene, and very likely arose from a different genomic locus.
Besides sequence differences, the intron–exon boundaries and structures are different for the AFGP genes from these two polar fishes (Figs. 2 and 3). Arctic cod AFGP gene contains three exons and two small introns—intron 1 (82 nt) intervenes in the 5′ UTR and intron 2 (69 nt) separates the translational start (Met codon) from the rest of the signal peptide (Figs. 2 and 3). Notothenioid AFGP genes contain a single large intron (1.9 kbp) with the same splice junctions as the first intron of the trypsinogen gene, which separates the small exon 1 (most of signal peptide) from the large exon 2 (remainder of signal peptide and AFGP polyprotein) (ref. 12; Figs. 2 and 3). Intron–exon boundaries generally remain identical among homologous genes, even in widely divergent organisms, as exemplified by the highly conserved three exons/two introns structure of insulin genes from hagfish to human (20). The boundaries of the six exons and five introns of the trypsinogen gene of the Antarctic notothenioid D. mawsoni (12) are conserved in rat trypsinogen (21) except for the loss of the small intron 3 (117 nt), which linked exons 3 and 4 as one exon in rat. The distinct dissimilarity of the intron–exon organizations of the Arctic cod and notothenioid AFGP genes indicates the two genes are not homologous.
Within the AFGP polyprotein coding region, two significant differences, the spacers and the AFGP tripeptide coding sequences, argue strongly for different ancestry. The spacers of notothenioid AFGPs are the highly conserved three residues, Leu/Phe-Ile/Asn-Phe, cleavable by a chymotrypsin-like protease (12, 13). The most frequent spacer, Leu-Ile-Phe (50%, 10/20 in Dm3L), is encoded as ttgattttt (Fig. 2B). The notothenioid spacer sequences probably adjoined the primordial AFGP sequence, and the two became amplified together to give rise to the polyprotein structure. By contrast, the spacers in Arctic cod AFGPs are Arg, which require a different protease, presumably trypsin-like. These Arg are almost exclusively encoded by aga (95%, 20/21 in Bs3–1) and always replace a Thr in the tripeptide repeats, and thus very likely arose from the mutation of a Thr of one of the repeats by a 1-nt transversion of aca to aga (45% of Thr encoded by aca in Bs3–1), and became a cleavage site. Not only are the origins of the spacers of the AFGP genes from the two fishes very likely different, the sequences of the spacers are so distinct it is inconceivable that one could be converted to the other through divergence of a common ancestral element. Nor is it logical that the processing (the protease involved) of the AFGP polyproteins of the two fish should be reinvented once they had come from a common ancestral molecule.
The highly repetitive tripeptide coding sequences in AFGP polyprotein genes suggests that extensive duplications of a primordial short sequence gave rise to both groups of AFGPs. If the duplication events were sufficiently recent, one can expect the nucleotide sequence of the duplicants to closely resemble that of the ancestral short sequence. This is indeed the case for the notothenioid AFGPs. Striking sequence similarity exists between the appropriately located candidate ancestral 9-nt coding element in trypsinogen gene, aca(Thr)-gcg(Ala)-gca(Ala) (12), and the coding sequences of the tripeptide repeats in AFGP genes, which, for the Dm3L AFGP gene, have the composition of aca(85% of Thr)-gcg/gct(39%/55% of Ala)-gca(95% of Ala). A 1-nt substitution at the first Ala codon, from gct to cct, would give rise to the substitution of the first Ala by Pro seen in the protein. The Pro codons in the Thr-Pro-Ala repeats in Dm3L AFGP gene are almost exclusively cct (97%). The same patterns of sequence similarities are seen in other notothenioid AFGP polyprotein genes (12, 13).
By contrast, the repetitive tripeptide coding sequences in the Arctic cod Bs3–1 AFGP gene have a very different composition, aca/act(45%/42% of Thr)-gca/gcg(51%/30% of Ala)-gca/gcc(53%/37% of Ala). One of the codons at each of the three amino acid positions (underlined) is either rarely or never used for the corresponding amino acid in notothenioid AFGP genes. The substitution of the first Ala by Pro likely resulted from a 1-nt substitution, changing gca/gcg to cca/ccg, as indicated by the frequency and composition of the Pro codons (cca/ccg, 50%/48%) in the Thr-Pro-Ala repeats encoded in the gene. None of the Pro is encoded by cct, which is the exclusive Pro codon in notothenioid AFGP genes (97% in Dm3L). The roughly equal usage of two particular codons for each of the three residues in the Thr-Ala/Pro-Ala repeats suggests that the primordial AFGP short sequence that became amplified might have consisted of two tripeptide repeats. Thus, besides the lack of evolutionary relatedness of Arctic cod AFGP gene to trypsinogen and notothenioid AFGP genes based on complete sequence dissimilarities in the signal peptide and UTR coding regions, the marked sequence differences within the AFGP coding regions of the two fishes provide additional support for separate genomic origin.
It could be argued that the extent of sequence dissimilarity observed between Antarctic notothenioid and Arctic cod AFGP genes is possible if they had diverged from a common ancestral AFGP sequence a very long time ago. However, this argument is not supported by molecular evidence or paleoclimatic data of the polar regions, which indicate that the evolution of AFGPs in both groups of fishes are recent and separate events. In the case of notothenioids, the very small amount of sequence divergence (4–7%) between AFGP and trypsinogen genes (especially in the intron sequences (7%), which are under no constraint to be conserved) translates into a divergence time of about 5–14 million years (12). Evolution of AFGPs during this time frame coincides remarkably well with the estimated mid-Miocene time frame [10–14 million years ago (mya)], during which the Antarctic Ocean cooled to freezing temperatures based on oxygen isotopic ratios of planktonic sediments (22–24). It also coincides well with the placement of the main phyletic radiation of the five AFGP-bearing Antarctic notothenioid families at about 7–15 mya, based on mitochondrial rRNA gene sequences (25). Although the time of AFGP evolution in the northern cods cannot be estimated by the rate of sequence divergence because its genomic origin is currently unidentified, paleoclimatic data indicate that the driving force for its AFGP evolution (i.e., the glaciation of the northern hemisphere and the freezing of the North Atlantic and North Pacific oceans) occurred much later, at about late Pliocene (2.5 mya) (26, 27), indicating that AFGP evolution in boreal cods was a more recent and separate event.
Before the glaciation of the polar regions, antifreeze protein would not be needed for survival in the ancestral stocks of the Antarctic notothenioids and northern cods. The divergence time of notothenioids and gadoids (cods) is not clear, but based on the gadiform fossils (28) and a notothenioid skull (29) identified from the same late Eocene (40 mya) deposits on Seymour Island in the Antarctic Peninsula (30), they diverged at least 40 mya, at a time when the Antarctic Ocean was significantly above freezing (5–7°C) (22–24), and the world’s oceans were much warmer (23). In addition, it has been widely accepted that the two fish taxa bear morphological differences at higher taxonomic levels and thus do not share a recent common ancestor (10, 31). Gadiforms are in fact believed to have evolved in the Northern Hemisphere (32), whereas notothenioids evolved in the Southern Ocean, and the modern notothenioid fauna is endemic to the Antarctic regions with no northern representatives (33, 34). The morphological, paleontological, and paleoclimatic evidence are consistent with the two fish taxa having diverged before antifreeze protein was needed, and thus they would not have inherited a common ancestral AFGP sequence and must have evolved their respective AFGPs independently.
The molecular evidence from the AFGP genes of the Antarctic notothenioids and Arctic cod in this study, in conjunction with other supporting evidence, argues strongly for convergent evolution of AFGPs from independent genomic loci and at different paleo time points. The similar polyprotein AFGP gene structures of these two unrelated polar fishes arose not from the descent from a common polyprotein progenitor gene, but from the tendency for short repetitive sequences to undergo expansion through slippage replication (35) and unequal crossing-over (36), especially under strong selectional pressure provided by the subzero polar oceans. However, the selection of an appropriate permutation of three codons that gave rise to the same mature glycotripeptide gene product capable of ice binding might not be fortuitous, but the selection was likely shaped by the structural specificity required for antifreeze ice interaction to take place. Antifreeze adsorption to ice involves a matching between the repeat spacing of hydrogen bonding moieties of the antifreeze molecule and the periodicity of water molecules in the ice lattice (4, 37). AFGPs have been found to adsorb to very specific faces (the prism planes) and align in specific directions (parallel to a axes) in ice crystals through a lattice match between specific hydroxyls in the disaccharides and the water molecules within these ice crystallographic parameters (37). The regular tripeptide-repeating structure might be required for the AFGPs to be properly glycosylated and to position the disaccharides appropriately for ice binding to occur.
Acknowledgments
This work was supported in part by National Science Foundation Office of Polar Programs Grants 90–19881 and 93–17629 to A.L.D. We also thank Dr. E. K. Zachariassen for his assistance in collecting Arctic cod specimens.
ABBREVIATIONS
- AFGP
antifreeze glycoprotein
- mya
million years ago
- RACE
rapid amplification of cDNA ends
- UTR
untranslated region.
Footnotes
References
- 1.DeVries A L. Science. 1971;172:1152–1155. doi: 10.1126/science.172.3988.1152. [DOI] [PubMed] [Google Scholar]
- 2.DeVries A L. Comp Biochem Physiol. 1982;A73:627–640. [Google Scholar]
- 3.Davies P L, Hew C L. FASEB J. 1990;4:2460–2468. doi: 10.1096/fasebj.4.8.2185972. [DOI] [PubMed] [Google Scholar]
- 4.Cheng C-H C, DeVries A L. In: Life Under Extreme Conditions. di Prisco G, editor. Berlin: Springer; 1991. pp. 1–14. [Google Scholar]
- 5.Hew C L, Joshi S, Wang N-C, Cao M-H, Ananthanarayanan V S. Eur J Biochem. 1985;151:167–172. doi: 10.1111/j.1432-1033.1985.tb09081.x. [DOI] [PubMed] [Google Scholar]
- 6.Ng N F, Hew C L. J Biol Chem. 1992;267:16069–16075. [PubMed] [Google Scholar]
- 7.Ewart K V, Fletcher G L. Mol Mar Biol Biotechnol. 1993;2:20–27. [PubMed] [Google Scholar]
- 8.O’Grady S M, Schrag J D, Raymond J A, DeVries A L. J Exp Zool. 1982;224:177–185. [Google Scholar]
- 9.Osuga D T, Feeney R E. J Biol Chem. 1978;253:5338–5343. [PubMed] [Google Scholar]
- 10.Nelson J S. Fishes of the World. New York: Wiley; 1994. [Google Scholar]
- 11.Fletcher G L, Hew C L, Joshi S B. J Can Zool. 1982;60:348–355. [Google Scholar]
- 12.Chen L, DeVries A L, Cheng C-H C. Proc Natl Acad Sci USA. 1997;94:3811–3816. doi: 10.1073/pnas.94.8.3811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hsiao K C, Cheng C-H C, Fernandes I E, Detrich H W, DeVries A L. Proc Natl Acad Sci USA. 1990;87:9265–9269. doi: 10.1073/pnas.87.23.9265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 15.Morris H R, Thompson M R, Osuga D T, Ahmed A I, Chan S M, Vandenheede J R, Feeney R E. J Biol Chem. 1978;253:5155–5162. [PubMed] [Google Scholar]
- 16.Ahlgren J A, DeVries A L. Polar Biol. 1984;3:93–97. [Google Scholar]
- 17.Cheng C-H C. In: Gene Expression and Manipulation in Aquatic Organisms. Ennion S, Goldspink G, editors. Cambridge, UK: Cambridge Univ. Press; 1995. pp. 1–20. [Google Scholar]
- 18.Scott G K, Fletcher G L, Davies P L. Can J Fish Aquat Sci. 1986;43:1028–1034. [Google Scholar]
- 19.Gudmundsdóttir Á, Gudmundsdóttir E, Óskarsson S, Bjarnason J B, Eakin A K, Craik S. Eur J Biochem. 1993;217:1091–1097. doi: 10.1111/j.1432-1033.1993.tb18341.x. [DOI] [PubMed] [Google Scholar]
- 20.Steiner D F, Chan S J, Welsh J M, Kwok S C M. Ann Rev Genet. 1985;19:463–484. doi: 10.1146/annurev.ge.19.120185.002335. [DOI] [PubMed] [Google Scholar]
- 21.Craik C S, Choo Q L, Swift G H, Quinto C, MacDonald R J, Rutter J R. J Biol Chem. 1984;259:14255–14264. [PubMed] [Google Scholar]
- 22.Kennett J P. J Geophys Res. 1977;82:3843–3860. [Google Scholar]
- 23.Kennett J P. Marine Geology. Englewood Cliffs, NJ: Prentice–Hall; 1982. [Google Scholar]
- 24.Clarke A. In: Antarctic Ecosystems: Ecological Change and Conservation. Kerry K R, Hempel G, editors. Berlin: Springer; 1990. pp. 9–22. [Google Scholar]
- 25.Bargelloni L, Ritchie P A, Patarnello T, Battaglia B, Lambert D M, Meyer A. Mol Biol Evol. 1994;11:854–863. doi: 10.1093/oxfordjournals.molbev.a040168. [DOI] [PubMed] [Google Scholar]
- 26.Shackleton N J, Backman J, Zimmerman H, Kent D V, Hall M A, Roberts D G, Schnitker D, Baldauf J G, Desprairies A, Homrighausen R, Huddlestun P, Keene J B, Kaltenback A J, Krumsiek K A O, Morton A C, Murray J W, Westber-Smith J. Nature (London) 1984;307:620–623. [Google Scholar]
- 27.Rea D K, Schrader H. Palaeogeogr Palaeoclimatol Palaeoecol. 1985;49:313–325. [Google Scholar]
- 28.Jerzmańska A. Pol Polar Res. 1988;9:421–435. [Google Scholar]
- 29.Eastman J T, Grande L. Antarct Sci. 1991;3:87–93. [Google Scholar]
- 30.Balushkin A V. J Ichthyol. 1995;34:10–23. [Google Scholar]
- 31.Patterson C, Rosen D E. Nat Hist Mus Los Angeles Co Sci Ser. 1989;32:5–36. [Google Scholar]
- 32.Svetovidov A N. Gadiformes. Jerusalem: Israel Program for Scientific Translation; 1948. ; trans. Monson, S. (Russian). [Google Scholar]
- 33.Eastman J T. Antarctic Fish Biology. San Diego: Academic; 1993. [Google Scholar]
- 34.Eastman J T. Am Zool. 1991;31:93–109. [Google Scholar]
- 35.Dover G A, Tautz D. Philos Trans R Soc Lond Ser B. 1986;312:275–289. doi: 10.1098/rstb.1986.0007. [DOI] [PubMed] [Google Scholar]
- 36.Maeda N, Smithies O. Annu Rev Genet. 1986;20:81–108. doi: 10.1146/annurev.ge.20.120186.000501. [DOI] [PubMed] [Google Scholar]
- 37.Knight C A, Driggers E, DeVries A L. Biophys J. 1993;64:252–259. doi: 10.1016/S0006-3495(93)81361-4. [DOI] [PMC free article] [PubMed] [Google Scholar]