Abstract
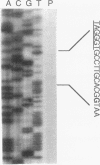
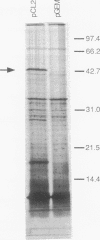
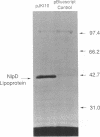
We report a 1.432-kb DNA sequence at 59 min on the Escherichia coli chromosome that connects the published sequences of the pcm gene for the isoaspartyl protein methyltransferase and that of the katF or rpoS (katF/rpoS) gene for a sigma factor involved in stationary-phase gene expression. Analysis of the DNA sequence reveals an open reading frame potentially encoding a polypeptide of 379 amino acids. The polypeptide sequence includes a consensus bacterial lipidation sequence present at residues 23 to 26 (Leu-Ala-Gly-Cys), four octapeptide proline- and glutamine-rich repeats of consensus sequence QQPQIQPV, and four heptapeptide threonine- and serine-rich repeats of consensus sequence PTA(S,T)TTE. The deduced amino acid sequence, especially in the C-terminal region, is similar to that of the Haemophilus somnus LppB lipoprotein outer membrane antigen (40% overall sequence identity; 77% identity in last 95 residues). The LppB lipoprotein binds Congo red dye and has been proposed to be a virulence determinant in H. somnus. Utilizing a plasmid construct with the E. coli gene under the control of a phage T7 promoter, we demonstrate the lipidation of this gene product by the incorporation of [3H]palmitic acid into a 42-kDa polypeptide. We also show that treatment of E. coli cells with globomycin, an inhibitor of the lipoprotein signal peptidase, results in the accumulation of a 46-kDa precursor. We thus designate the protein NlpD (new lipoprotein D). E. coli cells overexpressing NlpD bind Congo red dye, suggesting a common function with the H. somnus LppB protein. Disruption of the chromosomal E. coli nlpD gene by insertional mutagenesis results in decreased stationary-phase survival after 7 days.
Full text
PDF








Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barten D. M., O'Dea R. F. The function of protein carboxylmethyltransferase in eucaryotic cells. Life Sci. 1990;47(3):181–194. doi: 10.1016/0024-3205(90)90319-m. [DOI] [PubMed] [Google Scholar]
- Bouvier J., Pugsley A. P., Stragier P. A gene for a new lipoprotein in the dapA-purC interval of the Escherichia coli chromosome. J Bacteriol. 1991 Sep;173(17):5523–5531. doi: 10.1128/jb.173.17.5523-5531.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Båga M., Göransson M., Normark S., Uhlin B. E. Processed mRNA with differential stability in the regulation of E. coli pilin gene expression. Cell. 1988 Jan 29;52(2):197–206. doi: 10.1016/0092-8674(88)90508-9. [DOI] [PubMed] [Google Scholar]
- Chung C. T., Niemela S. L., Miller R. H. One-step preparation of competent Escherichia coli: transformation and storage of bacterial cells in the same solution. Proc Natl Acad Sci U S A. 1989 Apr;86(7):2172–2175. doi: 10.1073/pnas.86.7.2172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke S. Protein carboxyl methyltransferases: two distinct classes of enzymes. Annu Rev Biochem. 1985;54:479–506. doi: 10.1146/annurev.bi.54.070185.002403. [DOI] [PubMed] [Google Scholar]
- Corbeil L. B., Kania S. A., Gogolewski R. P. Characterization of immunodominant surface antigens of Haemophilus somnus. Infect Immun. 1991 Dec;59(12):4295–4301. doi: 10.1128/iai.59.12.4295-4301.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daskaleros P. A., Payne S. M. Congo red binding phenotype is associated with hemin binding and increased infectivity of Shigella flexneri in the HeLa cell model. Infect Immun. 1987 Jun;55(6):1393–1398. doi: 10.1128/iai.55.6.1393-1398.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang F. C., Libby S. J., Buchmeier N. A., Loewen P. C., Switala J., Harwood J., Guiney D. G. The alternative sigma factor katF (rpoS) regulates Salmonella virulence. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):11978–11982. doi: 10.1073/pnas.89.24.11978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich M. J., de Veaux L. C., Kadner R. J. Nucleotide sequence of the btuCED genes involved in vitamin B12 transport in Escherichia coli and homology with components of periplasmic-binding-protein-dependent transport systems. J Bacteriol. 1986 Sep;167(3):928–934. doi: 10.1128/jb.167.3.928-934.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J. C., Ding L., Clarke S. Purification, gene cloning, and sequence analysis of an L-isoaspartyl protein carboxyl methyltransferase from Escherichia coli. J Biol Chem. 1991 Aug 5;266(22):14562–14572. [PubMed] [Google Scholar]
- Galletti P., Ciardiello A., Ingrosso D., Di Donato A., D'Alessio G. Repair of isopeptide bonds by protein carboxyl O-methyltransferase: seminal ribonuclease as a model system. Biochemistry. 1988 Mar 8;27(5):1752–1757. doi: 10.1021/bi00405a055. [DOI] [PubMed] [Google Scholar]
- Gennity J. M., Inouye M. The protein sequence responsible for lipoprotein membrane localization in Escherichia coli exhibits remarkable specificity. J Biol Chem. 1991 Sep 5;266(25):16458–16464. [PubMed] [Google Scholar]
- Gennity J. M., Kim H., Inouye M. Structural determinants in addition to the amino-terminal sorting sequence influence membrane localization of Escherichia coli lipoproteins. J Bacteriol. 1992 Apr;174(7):2095–2101. doi: 10.1128/jb.174.7.2095-2101.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harley C. B., Reynolds R. P. Analysis of E. coli promoter sequences. Nucleic Acids Res. 1987 Mar 11;15(5):2343–2361. doi: 10.1093/nar/15.5.2343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S., Wu H. C. Lipoproteins in bacteria. J Bioenerg Biomembr. 1990 Jun;22(3):451–471. doi: 10.1007/BF00763177. [DOI] [PubMed] [Google Scholar]
- Hengge-Aronis R. Survival of hunger and stress: the role of rpoS in early stationary phase gene regulation in E. coli. Cell. 1993 Jan 29;72(2):165–168. doi: 10.1016/0092-8674(93)90655-a. [DOI] [PubMed] [Google Scholar]
- Ichihara S., Hussain M., Mizushima S. Characterization of new membrane lipoproteins and their precursors of Escherichia coli. J Biol Chem. 1981 Mar 25;256(6):3125–3129. [PubMed] [Google Scholar]
- Jarrett J. T., Lansbury P. T., Jr Amyloid fibril formation requires a chemically discriminating nucleation event: studies of an amyloidogenic sequence from the bacterial protein OsmB. Biochemistry. 1992 Dec 15;31(49):12345–12352. doi: 10.1021/bi00164a008. [DOI] [PubMed] [Google Scholar]
- Jin T., Rudd K. E., Inouye M. The nlpA lipoprotein gene is located near the selC tRNA gene on the Escherichia coli chromosome. J Bacteriol. 1992 Jun;174(11):3822–3823. doi: 10.1128/jb.174.11.3822-3823.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson B. A., Murray E. D., Jr, Clarke S., Glass D. B., Aswad D. W. Protein carboxyl methyltransferase facilitates conversion of atypical L-isoaspartyl peptides to normal L-aspartyl peptides. J Biol Chem. 1987 Apr 25;262(12):5622–5629. [PubMed] [Google Scholar]
- Jung J. U., Gutierrez C., Villarejo M. R. Sequence of an osmotically inducible lipoprotein gene. J Bacteriol. 1989 Jan;171(1):511–520. doi: 10.1128/jb.171.1.511-520.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kay W. W., Phipps B. M., Ishiguro E. E., Trust T. J. Porphyrin binding by the surface array virulence protein of Aeromonas salmonicida. J Bacteriol. 1985 Dec;164(3):1332–1336. doi: 10.1128/jb.164.3.1332-1336.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lal A. A., de la Cruz V. F., Welsh J. A., Charoenvit Y., Maloy W. L., McCutchan T. F. Structure of the gene encoding the circumsporozoite protein of Plasmodium yoelii. A rodent model for examining antimalarial sporozoite vaccines. J Biol Chem. 1987 Mar 5;262(7):2937–2940. [PubMed] [Google Scholar]
- Lange R., Hengge-Aronis R. Identification of a central regulator of stationary-phase gene expression in Escherichia coli. Mol Microbiol. 1991 Jan;5(1):49–59. doi: 10.1111/j.1365-2958.1991.tb01825.x. [DOI] [PubMed] [Google Scholar]
- Li C., Clarke S. A protein methyltransferase specific for altered aspartyl residues is important in Escherichia coli stationary-phase survival and heat-shock resistance. Proc Natl Acad Sci U S A. 1992 Oct 15;89(20):9885–9889. doi: 10.1073/pnas.89.20.9885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. J., Dougan G., Novotny P., Charles I. G. P.70 pertactin, an outer-membrane protein from Bordetella parapertussis: cloning, nucleotide sequence and surface expression in Escherichia coli. Mol Microbiol. 1991 Feb;5(2):409–417. doi: 10.1111/j.1365-2958.1991.tb02123.x. [DOI] [PubMed] [Google Scholar]
- Loewen P. C., von Ossowski I., Switala J., Mulvey M. R. KatF (sigma S) synthesis in Escherichia coli is subject to posttranscriptional regulation. J Bacteriol. 1993 Apr;175(7):2150–2153. doi: 10.1128/jb.175.7.2150-2153.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCann M. P., Fraley C. D., Matin A. The putative sigma factor KatF is regulated posttranscriptionally during carbon starvation. J Bacteriol. 1993 Apr;175(7):2143–2149. doi: 10.1128/jb.175.7.2143-2149.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden P. N., Clarke S. Conversion of isoaspartyl peptides to normal peptides: implications for the cellular repair of damaged proteins. Proc Natl Acad Sci U S A. 1987 May;84(9):2595–2599. doi: 10.1073/pnas.84.9.2595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. R., Loewen P. C. Nucleotide sequence of katF of Escherichia coli suggests KatF protein is a novel sigma transcription factor. Nucleic Acids Res. 1989 Dec 11;17(23):9979–9991. doi: 10.1093/nar/17.23.9979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulvey M. R., Sorby P. A., Triggs-Raine B. L., Loewen P. C. Cloning and physical characterization of katE and katF required for catalase HPII expression in Escherichia coli. Gene. 1988 Dec 20;73(2):337–345. doi: 10.1016/0378-1119(88)90498-2. [DOI] [PubMed] [Google Scholar]
- Mulvey M. R., Switala J., Borys A., Loewen P. C. Regulation of transcription of katE and katF in Escherichia coli. J Bacteriol. 1990 Dec;172(12):6713–6720. doi: 10.1128/jb.172.12.6713-6720.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Médigue C., Bouché J. P., Hénaut A., Danchin A. Mapping of sequenced genes (700 kbp) in the restriction map of the Escherichia coli chromosome. Mol Microbiol. 1990 Feb;4(2):169–187. doi: 10.1111/j.1365-2958.1990.tb00585.x. [DOI] [PubMed] [Google Scholar]
- Payne S. M., Finkelstein R. A. Detection and differentiation of iron-responsive avirulent mutants on Congo red agar. Infect Immun. 1977 Oct;18(1):94–98. doi: 10.1128/iai.18.1.94-98.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prpic J. K., Robins-Browne R. M., Davey R. B. Differentiation between virulent and avirulent Yersinia enterocolitica isolates by using Congo red agar. J Clin Microbiol. 1983 Sep;18(3):486–490. doi: 10.1128/jcm.18.3.486-490.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rupp W. D., Wilde C. E., 3rd, Reno D. L., Howard-Flanders P. Exchanges between DNA strands in ultraviolet-irradiated Escherichia coli. J Mol Biol. 1971 Oct 14;61(1):25–44. doi: 10.1016/0022-2836(71)90204-x. [DOI] [PubMed] [Google Scholar]
- Studier F. W., Rosenberg A. H., Dunn J. J., Dubendorff J. W. Use of T7 RNA polymerase to direct expression of cloned genes. Methods Enzymol. 1990;185:60–89. doi: 10.1016/0076-6879(90)85008-c. [DOI] [PubMed] [Google Scholar]
- Tabor S., Richardson C. C. A bacteriophage T7 RNA polymerase/promoter system for controlled exclusive expression of specific genes. Proc Natl Acad Sci U S A. 1985 Feb;82(4):1074–1078. doi: 10.1073/pnas.82.4.1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Takayanagi Y., Fujita N., Ishihama A., Takahashi H. Heterogeneity of the principal sigma factor in Escherichia coli: the rpoS gene product, sigma 38, is a second principal sigma factor of RNA polymerase in stationary-phase Escherichia coli. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3511–3515. doi: 10.1073/pnas.90.8.3511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Theisen M., Rioux C. R., Potter A. A. Molecular cloning, nucleotide sequence, and characterization of lppB, encoding an antigenic 40-kilodalton lipoprotein of Haemophilus somnus. Infect Immun. 1993 May;61(5):1793–1798. doi: 10.1128/iai.61.5.1793-1798.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokunaga M., Tokunaga H., Wu H. C. Post-translational modification and processing of Escherichia coli prolipoprotein in vitro. Proc Natl Acad Sci U S A. 1982 Apr;79(7):2255–2259. doi: 10.1073/pnas.79.7.2255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber J. L., Egan J. E., Lyon J. A., Wirtz R. A., Charoenvit Y., Maloy W. L., Hockmeyer W. T. Plasmodium berghei: cloning of the circumsporozoite protein gene. Exp Parasitol. 1987 Jun;63(3):295–300. doi: 10.1016/0014-4894(87)90176-7. [DOI] [PubMed] [Google Scholar]
- Yu F., Inouye S., Inouye M. Lipoprotein-28, a cytoplasmic membrane lipoprotein from Escherichia coli. Cloning, DNA sequence, and expression of its gene. J Biol Chem. 1986 Feb 15;261(5):2284–2288. [PubMed] [Google Scholar]
- von Heijne G. The structure of signal peptides from bacterial lipoproteins. Protein Eng. 1989 May;2(7):531–534. doi: 10.1093/protein/2.7.531. [DOI] [PubMed] [Google Scholar]