Abstract
Theory concerning the evolution of sex and recombination and mutation load relies on information on rates and distributions of effects of deleterious mutations. Direct information on the genomic mutation rate in Drosophila implies that an accumulation of mildly deleterious mutations reduces viability of populations by at least 1% per generation. We carried out an experiment to measure the deleterious mutation rate in Caenorhabditis elegans, in which independent sublines were maintained with one hermaphrodite parent per generation, conditions that minimize the opportunity for natural selection and lead to random fixation of deleterious mutations. After 60 generations of mutation accumulation, negligible changes in mean reproductive output and lifespan occurred, but the genetic variance increased at rates typical for life history traits in other species. The estimated deleterious mutation rate per haploid genome for fitness, U, was 0.0026, a figure two orders of magnitude smaller than previously measured for viability in Drosophila.
Despite its importance for evolutionary theory concerning the evolution of sex and recombination (1, 2), maintenance of variation (3, 4), and mutation load (5–7), direct information on rates and distributions of effects of deleterious mutations in eukaryotes comes exclusively from experiments in Drosophila in which replicated second chromosomes are allowed to accumulate spontaneous mutations sheltered from selection by a balancer chromosome (8–10). These experiments are widely interpreted as demonstrating that mildly deleterious mutations occur at the rate of ≈1 event per diploid genome per generation (i.e., at least 20 times more frequently than lethals), with average effects on viability of 1–2% (5–7). Indirect estimates of deleterious mutation rates in naturally self-fertilizing plants based on relative fitnesses of selfed and outbred progeny also imply a deleterious mutation rate of ≈1 (11) but rely on the assumption that populations are at mutation–selection balance (12). The estimated mutation rates and mean effects frequently are assumed in population genetics and evolutionary models (1–5, 13–15). For example, the deterministic mutation hypothesis for the evolution of sex depends on a deleterious mutation rate greater than 1 (1), but the ability of small populations to persist, faced by such an accumulation of deleterious mutations, is problematical (13–15).
The primary aim of this study was to estimate the rate of change of mean and variance of lifetime reproductive output due to the fixation of newly arising mutations in Caenorhabditis elegans. The experiment also allowed an estimate for U (estimated deleterious mutation rate per haploid genome for fitness) to be obtained and inferences to be made on the nature of the distribution of mutation effects (16). Spontaneous mutations were allowed to accumulate in 50 independent lines derived from an inbred progenitor maintained for 60 generations by transfer of single, randomly picked hermaphrodite larvae. A self-fertilized parent was therefore the sole contributor to the next generation. By maintaining the lines with only one parent per generation, competition among larvae was kept to a minimum. Furthermore, because the effective population size for each mutation accumulation (MA) line is 1, random sampling of gametes dominates selection among offspring. Fates of new mutations, with the exception of those with lethal or nearly lethal effects, depend largely on chance (Fig. 1). For example, mutations with a selective disadvantage in the homozygote of 50% are expected to be fixed two–thirds as frequently as neutrals; mildly deleterious mutations (with effects, say, <10%) become fixed at essentially the same rate as neutrals. Thus, the experimental design does not allow measurement of the rate of lethal or severely deleterious mutations but focuses on the overall effect on fitness traits of fixation of the more frequent class of mutation thought to be responsible for the bulk of mutation load in Drosophila (5–7).
Figure 1.
Fixation probability of a new mutation occurring in a self-fertilizing line plotted against its selection coefficient s. The fitnesses of the wild-type, heterozygote, and mutant genotypes were 1, 1 − hs, and 1 − s, respectively. For the case of selfing, it can be shown that the function relating fixation probability (u) and s is u = (1 − s)/(2 − s), and the mean time to fixation or loss is three generations. Fixation probability is independent of h, the degree of dominance of the mutation.
By taking advantage of the fact that C. elegans can be kept frozen at −70°C, we carried out assays for reproductive output and lifespan in the MA lines contemporaneously for worms from several generations. The productivity trait measured was closely related to fitness because it included viability of the parents after the L3 stage, fertility of the parents, and viability of their offspring up to the age at which they were counted (L3: young adult stage). We also measured mutational effects on lifespan, a life history trait potentially related to fitness. Both traits show high levels of genetic variation between laboratory populations derived from separate wild isolates (17) although the level of variability within natural populations is unknown.
MATERIALS AND METHODS
Strains and Culture Conditions.
A wild-type N2 strain of C. elegans was the progenitor strain for the MA experiment. The MA lines were maintained and life history traits were assayed on MYOB Agar plates (18). The food source for the worms was a 25-μl spot of a suspension of Escherichia coli strain OP50 allowed to grow overnight (19). During the 7 months of the MA phase of the experiment, cultures were maintained at 25°C; because of ambient conditions, occasional temperature fluctuations to 26°C occurred between generations 25 and 45.
Mutation Accumulation.
From a single inbred progenitor (generation 0), 50 MA lines of a single hermaphrodite parent each were maintained for 60 generations by twice-weekly transfer of single L3 larvae picked at random from a position near the edge of the worms’ food. In cases in which a parental worm failed to produce progeny (1 in 14, on average), an L3 larva was substituted from the plate from the previous generation. Competition among progeny would potentially be more severe in these cases, but 85% of mutants never segregate in such conditions (with N = 1, most mutants are fixed or lost in three generations). Furthermore, competition would not affect fates of mildly deleterious mutations because fates of mutants are dominated by random sampling unless their selective values approach 1 (Fig. 1). At generations 0, 32, and 60, cultures were cryopreserved (19) and kept at −70°C until the lifetime reproductive output and lifespan assays.
Reproductive Output Assay.
Before the assay, after thawing, 48 MA lines from generation 60 (two lines were lost during the MA phase), 34 lines from generation 32, and 40 replicates from generation 0 were maintained synchronously at 20°C for three generations. A complete set was not available from generation 32 because of a freezer fault. Productivity was assayed contemporaneously for all mutation accumulation lines. For each line, four hermaphrodites were allowed to lay eggs on a plate for ≈2 h and then were removed. After 44 h, hermaphrodite progeny, which were at the L3 stage, were transferred to plates in pairs and transferred 48 h later and at 24-h intervals for the duration of their reproductive period. The daily progeny were counted from hour 46 after transfer of their parents and were in the range of 46–70 h post egg lay. To prevent further growth, plates were kept at 4°C until the time of counting. For each line, there were two replicates in the assay, and the whole assay was performed twice (i.e., a total of eight parents were assayed per line). Counting was performed manually with the plates cooled to inhibit the worms from moving. The correlation between counts made as above and counts made by hand-picking was 99% (n = 27). Plates were randomized and randomly allocated between two measurers, and counters were blinded to line or generation identities. A total of 248,992 progeny was counted.
Lifespan Assay.
The parents for the productivity assay also were used in a lifespan assay, which was performed at 20°C throughout, with a design similar to that described by Johnson and Wood (20). For each line, there were two plates containing two worms transferred daily during their reproductive period. After the reproductive period, the replicates were combined and worms were scored 5–7 times per week and transferred 2–3 times per week. The whole assay was replicated twice, i.e., the lifespans of eight worms per line were measured. Worms observed not to be moving were scored as dead if they failed to respond to touch, showed a lack of osmotic turgor, or showed visible signs of decay (20).
Estimation of Between-Line Variance.
An ANOVA was used to estimate the between-line component of variance by partitioning the variance between and within lines. Additional effects fitted in the analysis were measurer (two people carried out the assay) and assay number (two levels). Measurer was nonsignificant in each generation for both productivity and lifespan. A significant assay number effect was detected in generation 32 for both productivity (P = 0.004) and lifespan (P = 0.03), but the effect was nonsignificant for data from generations 0 and 60.
Maximum Likelihood (ML) Estimation of U and Inference of the Distribution of Mutation Effects.
The observed distribution of estimated productivities (or lifespans) of the MA lines from different generations provides information on the underlying genomic mutation rate and distribution of mutation effects. However, the lack of prior knowledge of the form of the distribution of mutation effects requires that some distribution be assumed and its parameters estimated in the model. A γ distribution was chosen for modeling purposes because it can take a wide variety of shapes (21). The density function for the γ distribution of effects, a, is:
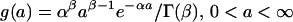 |
where α and β are scale and shape parameters, respectively, and are estimated, and Γ(.) is the γ function. In principle, it is possible to use data from several generations in the ML analysis (16), but for reasons of computational feasibility, the analysis was restricted to data from generations 0 and 60 only, which should be independent and most informative. Individual observed values, Zi, were assumed to be the sum of effects of an unknown number of mutation effects, a normally distributed environmental effect, plus the overall population mean. The number of mutation events per generation was assumed to be Poisson distributed with parameter U. The likelihood, L, of an observation Zi from generation t was computed according to an additive genetic model,
 |
where ƒ(x) is the normal density function, p(y|z) is the Poisson distribution function for y mutation events given an expected number z, and μ and σE2 are the population mean and residual variance, respectively, estimated as parameters. To allow for the possibility of some mutations increasing fitness, the γ distribution was reflected at 0, with proportions π1 and 1 − π1 of mutations increasing and decreasing productivity, respectively, and π1 also was estimated as a parameter in the likelihood evaluation. Monte Carlo integration was used to compute the likelihood of the data as a function of the model parameters (U, α, β, μ, σE2, π1), and the overall log likelihood (ΣlnL) was the sum of lnL for the independent observations (16). Evaluation by numerical integration also is possible, and gives essentially the same results as the Monte Carlo method (P.D.K., unpublished work). Approximate confidence limits were inferred by computing profile likelihoods with respect to individual parameters, with 95% confidence limits corresponding to a drop in ΣlnL of 2 from the maximum. Estimation of U and mutation distribution parameters by ML is superior to the method of inference used in classic MA experiments in Drosophila [which relies on information in the change of mean and between-line variance only (8–10)] because ML uses all of the information available in the distribution of phenotypic values of the lines.
RESULTS
Changes of Mean and Variance of Lifetime Productivity.
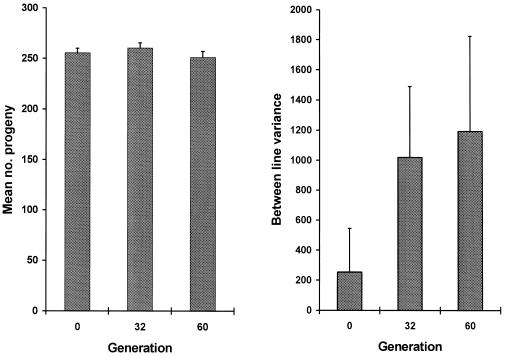
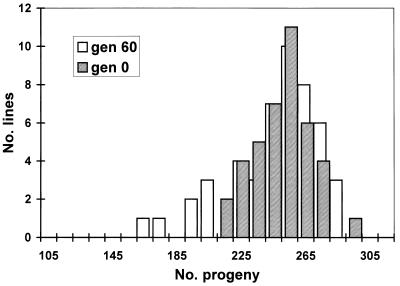
As expected, an accumulation of spontaneous mutations led to an increase in the among-line variance of the number of progeny produced (Fig. 2). However, the change in mean productivity over the 60 generations was very small. The estimated rate of change of productivity was −0.030% per generation (SE 0.03%). An ANOVA was used to estimate the between-line variance component VL. The increment in variance per generation from mutation, VM, is related to VL according to VL ≃ 2tVM, where t is the generation number (22). VL was nonsignificant for generation 0 (P > 0.1) but significant for both generations 32 and 60 (P < 0.01). The VM estimate obtained by a weighted regression of VL on generation number was 1.2 × 10−3 of the environmental variance (bootstrap SE 0.8 × 10−3). Therefore, the heritability increased by 0.12% per generation from mutation, a typical figure for life history traits (23). The mutational coefficient of variation, CVM = 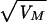 /mean productivity, was 1.2%, also a typical figure (23). The distribution of progeny number at generation 60 (Fig. 3) shows that the increase in variance was caused largely by a few lines, presumably carrying major mutations reducing fitness, and the majority of MA lines had productivities within the range for generation 0. With the experimental design used here, mutations with lethal or nearly lethal effects were not maintained in the lines (Fig. 1).
/mean productivity, was 1.2%, also a typical figure (23). The distribution of progeny number at generation 60 (Fig. 3) shows that the increase in variance was caused largely by a few lines, presumably carrying major mutations reducing fitness, and the majority of MA lines had productivities within the range for generation 0. With the experimental design used here, mutations with lethal or nearly lethal effects were not maintained in the lines (Fig. 1).
Figure 2.
Mean number of worms produced per hermaphrodite in the MA lines and the between-line component of variance of productivity estimated by ANOVA. (Bar = 1 SE.)
Figure 3.
Distributions of mean productivities of 48 mutation accumulation lines from generation 60 and 40 replicates from generation 0. gen, generation.
Genomic Mutation Rate and Distribution of Mutation Effects for Productivity.
The haploid genomic mutation rate per gamete per generation, U, was estimated by ML. The analysis gave an estimate for U of 0.0026, with approximate 95% confidence limits of ≈0.001 and 0.01, respectively. The mean mutational effect on productivity was estimated to be 21% (with 95% confidence limits of 6% and 29%). Effects on productivity were inferred to be exclusively negative although the upper limit for the proportion of mutations increasing productivity was 28%. A γ distribution of mutation effects with a very high value for β (i.e, a distribution for which all effects were approximately equal) gave the best fit to the data; γ distributions more leptokurtic than an exponential distribution (β < 1) were found to be significantly less likely than the ML distribution. An estimate of the number of lines containing a mutation is therefore 2 × 0.0026 (estimated diploid mutation rate) × 48 (surviving lines) × 60 (generations) × 1/2 (probability of fixation) ≃ 7, a figure that agrees well with the appearance of the distribution of productivities of the lines at generation 60 (Fig. 3). The predicted change in mean productivity is 2 × 0.0026 × 60 × 1/2 × 21% (mean mutant effect) ≃ 3%, which is similar to the observed change (2%; Fig. 2). Thus, the results of the analysis imply the occurrence of few mutations of large effect decreasing productivity (but mutations of small effect were not detected) and is consistent with the observation of an increase in variance between lines with little decrease in mean productivity (Fig. 2). A model with a mixed distribution of mutation effects in which a fraction of effects are equal and the remaining fraction have effects sampled from a γ distribution (24) did not give a significant increase in likelihood. Comparison of the numbers of progeny produced on different days suggested that mutation accumulation led to slightly reduced productivities in days 1 and 2 of the assay, but the reduction was partially offset by increased productivity in day 3. The overall effect was small, however, because ≈80% of progeny were produced in days 1 and 2 (data not shown).
An alternative method to infer the genomic mutation rate was devised by Bateman (25), in which mutation effects are assumed to be equal, and U is equated to the ratio of the square of the change of mean fitness to the change of variance, U = M2/V (9). Application of this formula gives an estimate for U of 0.0762/8.89 = 0.00065 and an estimate of the mean mutation effect of V/M = 117 worms = 46%.
Lifespan.
To investigate the effects of random fixation of spontaneous mutations on lifespan, we measured lifespan of worms from the MA lines and from the generation 0 replicates. The life expectancy curve showed only minor changes in shape after 60 generations, and mean lifespan was reduced only slightly (Fig. 4). The genetic variance for lifespan increased at the rate per generation of 4 × 10−4 of the environmental variance of the trait (CVM = 0.5%), but for this case the increase in variance was nonsignificant. A minimum estimate for the haploid genomic mutation rate for lifespan obtained by maximum likelihood was 0.0015.
Figure 4.
Proportions of worms surviving each day for the MA lines from generations 0, 32, and 60. Mean lifespan for generations 0, 32, and 60 was 14.0, 14.0, and 13.4 days, respectively. Gen, generation.
DISCUSSION
The principal finding of this study is a very small decrease in mean reproductive output (≈0.03% per generation) from the random accumulation of spontaneous mutations, but heritable variation from mutation increased at a rate similar to that observed previously for life history traits in other species (23). The negligible change of mean reproductive output in C. elegans contrasts sharply with the large reductions of mean viability seen in mutation accumulation experiments in Drosophila (8–10). The Drosophila experiments involved the accumulation of mutations on replicated, intact second chromosomes sheltered from natural selection by balancer chromosomes. Mean viability was observed to decline by 1–2% per generation relative to the balancer when extrapolated for the whole Drosophila genome. In common with all mutation accumulation experiments, including the present one, viability and fertility selection would not have been completely eliminated. A large part of the drop in viability was inferred to be due to the accumulation of mildly deleterious mutations with effects of 1–2% occurring at the rate of ≈0.5 events per haploid genome per generation (5–7). Our analysis shows that such a large class of mildly deleterious mutations is essentially absent in C. elegans or that their effects are so small as to make their presence undetectable. This result is important because the consequence of deleterious mutation for long term survival of populations is very sensitive to the fraction of mutations with small effects. For constant U × average effect, vulnerability of populations to deleterious mutation accumulation is greatest with small average effects and high U because fates of mutant alleles with small effects are most strongly influenced by genetic drift (14, 15).
In E. coli, a minimum estimate for the deleterious mutation rate for fitness of 0.0002 recently has been obtained (26). By accounting for the difference in genome size and the number of germ line divisions between E. coli and Drosophila, an adjusted estimate for U in E. coli corresponds closely to the minimum estimate of the genomic mutation rate for viability in Drosophila (≈0.3; refs. 5–7). If the C. elegans data are analyzed by the same Mukai–Bateman technique as above (9, 25), our unadjusted minimum estimate for U is 0.0006, i.e., ≈3 times higher than the minimum estimate for E. coli. The adjusted figure is 100 times smaller than the minimum estimated rate for Drosophila, i.e., 0.0006 (estimated U) × 1.7 (ratio between Drosphilia melanogaster and C. elegans genome sizes) × 3 (approximate ratio between Drosophila and C. elegans number of germ line divisions) ≃ 0.003.
The C. elegans genome contains ≈1.7 times fewer base pairs than Drosophila (27, 28), but the species have similar numbers of genes; current estimates are in the ranges 13,100–17,800 and 12,000–16,000 respectively (29, 30). Estimates of their per locus mutation rates also are similar (27, 31). Furthermore, measured lethal mutation rates differ by only a factor of 3; spontaneous lethal mutations accumulate at the rate of ≈0.01 in Drosophila (7), and in C. elegans the corresponding rate is ≈0.004 (32). A fundamental difference in underlying mutation rates is not, therefore, a plausible explanation for the difference in detectable genomic mutation rates for life history traits between the two species. There are at least three possible explanations that could account for the different mutation rate estimates. First, in the Drosophila experiments involving balancers, the trait measured was competitive viability. A competitive environment is likely to reveal greater differences between genotypes than a noncompetitive assay, as used here (33) [although “quasinormal” chromosomes in the experiments involving balancers showed surprisingly little genetic variance (24)]. However, mutational variation in Drosophila for major fitness components assayed noncompetitively accrues at as high a rate as for competitive viability (23, 34). Also, the contribution to the deleterious mutation load from ethyl methanesulfonate-induced mutations affecting fertility and other life history traits assayed noncompetitively may actually be higher than the contribution from competitive viability (7). A second explanation is that Drosophila could suffer from classes of mutation event, for example transposable element insertions, that occur at a high rate and have mildly deleterious effects. Artificially induced P-element insertions in autosomes of Drosophila have large average effects on viability (35) but effects of only ≈1% when inserted into the X chromosome (36), and it is possible that other classes of element also generate mutations mostly with small fitness effects. Such an explanation would imply that estimates of U from Drosophila are not generally applicable because transposition rates vary widely (37, 38). Finally, the large apparent reductions in mean viability of Drosophila chromosomes in mutation accumulation experiments involving balancers were accompanied by unexpectedly small increases in the between-line variance, raising the possibility that the changes of mean viability were artifacts (24). Evolution of the balancer system fueled by variation from gene conversion appearing de novo in the balancer stock is a possible mechanism.
The paradigm of a high rate for mildly deleterious mutations from chromosome balancer experiments in Drosophila has posed a major theoretical problem for the long term survival of populations, particularly for species with low normal reproductive capacities, like humans (5–7, 13–15). In C. elegans, we have measured a deleterious mutation rate at least 100 times smaller than previously assumed, and we did not detect minor deleterious mutations. Our estimated average mutation effect was 10 times larger that that for the Drosophila balancer studies. Our findings are consistent with recent observations of small changes in fitness traits in long term Drosophila mutation accumulation experiments not involving balancers (34) and with the persistence of some small populations.
Acknowledgments
We thank P. C. Phillips and W. G. Hill for helpful advice, and N. H. Barton, B. Charlesworth, E. Davies, and M. Lynch for constructive comments on the manuscript. This work was supported by the Royal Society (P.D.K.) and the Biotechnology and Biological Sciences Research Council (A.C.).
ABBREVIATIONS
- MA
mutation accumulation
- ML
maximum likelihood
References
- 1.Kondrashov A S. Nature (London) 1988;336:435–440. doi: 10.1038/336435a0. [DOI] [PubMed] [Google Scholar]
- 2.Charlesworth B. Curr Biol. 1996;6:149–162. doi: 10.1016/s0960-9822(02)00448-7. [DOI] [PubMed] [Google Scholar]
- 3.Barton N H, Turelli M. Annu Rev Genet. 1989;23:337–370. doi: 10.1146/annurev.ge.23.120189.002005. [DOI] [PubMed] [Google Scholar]
- 4.Nordborg M, Charlesworth B, Charlesworth D. Genet Res. 1996;67:159–174. doi: 10.1017/s0016672300033619. [DOI] [PubMed] [Google Scholar]
- 5.Crow J F. Oxford Surv Evol Biol. 1993;9:3–42. [Google Scholar]
- 6.Simmons M J, Crow J F. Annu Rev Genet. 1977;11:49–78. doi: 10.1146/annurev.ge.11.120177.000405. [DOI] [PubMed] [Google Scholar]
- 7.Crow J F, Simmons M J. In: The Genetics and Biology of Drosophila. Ashburner M, Carson H L, Thompson J N, editors. 3C. London: Academic; 1983. pp. 1–35. [Google Scholar]
- 8.Mukai T. Genetics. 1964;50:1–19. doi: 10.1093/genetics/50.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mukai T, Chigusa S I, Mettler L E, Crow J F. Genetics. 1972;72:333–355. doi: 10.1093/genetics/72.2.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ohnishi O. Genetics. 1977;87:529–545. doi: 10.1093/genetics/87.3.529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnston M O, Schoen D J. Science. 1995;267:226–229. doi: 10.1126/science.267.5195.226. [DOI] [PubMed] [Google Scholar]
- 12.Charlesworth B, Charlesworth D, Morgan M T. Nature (London) 1990;347:380–382. [Google Scholar]
- 13.Kondrashov A S. J Theor Biol. 1995;175:583–594. doi: 10.1006/jtbi.1995.0167. [DOI] [PubMed] [Google Scholar]
- 14.Lande R. Conserv Biol. 1995;9:782–791. [Google Scholar]
- 15.Lynch M, Conery J, Burger R. Am Nat. 1995;146:489–518. [Google Scholar]
- 16.Keightley P D. Genetics. 1994;138:1315–1322. doi: 10.1093/genetics/138.4.1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Johnson T E, Hutchinson E W. Genetics. 1993;134:465–474. doi: 10.1093/genetics/134.2.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lambie E J. Worm Breeder’s Gazette. 1994;13:12. [Google Scholar]
- 19.Sulston J, Hodgkin J. In: The Nematode Caenorhabditis elegans. Wood W B, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1988. [Google Scholar]
- 20.Johnson T E, Wood W B. Proc Natl Acad Sci USA. 1982;79:6603–6607. doi: 10.1073/pnas.79.21.6603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keightley P D, Hill W G. Genet Res. 1988;52:33–43. doi: 10.1017/s0016672300027282. [DOI] [PubMed] [Google Scholar]
- 22.Lynch M, Hill W G. Evolution. 1986;40:915–935. doi: 10.1111/j.1558-5646.1986.tb00561.x. [DOI] [PubMed] [Google Scholar]
- 23.Houle D, Morikawa B, Lynch M. Genetics. 1996;143:1467–1483. doi: 10.1093/genetics/143.3.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Keightley P D. Genetics. 1997;144:1993–1999. doi: 10.1093/genetics/144.4.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bateman A J. Int J Radiat Biol. 1959;1:170–180. [Google Scholar]
- 26.Kibota T T, Lynch M. Nature (London) 1996;381:694–696. doi: 10.1038/381694a0. [DOI] [PubMed] [Google Scholar]
- 27.Ashburner M. Drosophila: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 28.Coulson A, Huynh C, Kozono V, Shownkeen R. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism. Epstein H F, Shakes D C, editors. London: Academic; 1995. pp. 534–549. [Google Scholar]
- 29.Bird A P. Trends Genet. 1995;11:94–100. doi: 10.1016/S0168-9525(00)89009-5. [DOI] [PubMed] [Google Scholar]
- 30.Hodgkin J, Plasterk R H A, Waterston R H. Science. 1995;270:410–414. doi: 10.1126/science.270.5235.410. [DOI] [PubMed] [Google Scholar]
- 31.Anderson P. In: Caenorhabditis elegans: Modern Biological Analysis of an Organism. Epstein H F, Shakes D C, editors. London: Academic; 1995. pp. 31–54. [Google Scholar]
- 32.Rosenbluth R W, Cuddeford C, Baillie D L. Mutat Res. 1983;110:39–48. [Google Scholar]
- 33.Kondrashov A S, Houle D. Proc R Soc London. 1994;258:221–227. doi: 10.1098/rspb.1994.0166. [DOI] [PubMed] [Google Scholar]
- 34.Fernández J, López-Fanjul C. Genetics. 1996;143:829–837. doi: 10.1093/genetics/143.2.829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mackay T F C, Lyman R, Jackson M S. Genetics. 1992;130:315–332. doi: 10.1093/genetics/130.2.315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Eanes W F, Wesley C, Hey J, Houle D, Ajioka J W. Genet Res. 1988;52:17–26. [Google Scholar]
- 37.Pasyukova E G, Nuzhdin S V. Mol Gen Genet. 1993;240:302–306. doi: 10.1007/BF00277071. [DOI] [PubMed] [Google Scholar]
- 38.Charlesworth B, Jarne P, Assimacopoulos S. Genet Res. 1994;64:183–198. doi: 10.1017/s0016672300032845. [DOI] [PubMed] [Google Scholar]