Abstract
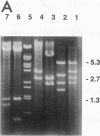
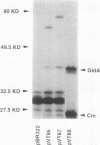
gldA, the structural gene for the NAD(+)-dependent glycerol dehydrogenase, was mapped at 89.2 min on the Escherichia coli linkage map, cotransducible with, but not adjacent to, the glpFKX operon encoding the proteins for the uptake and phosphorylation of glycerol. gldA was cloned, and its position on the physical map of E. coli was determined. The expression of gldA was induced by hydroxyacetone under stationary-phase growth conditions.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ASNIS R. E., BRODIE A. F. A glycerol dehydrogenase from Escherichia coli. J Biol Chem. 1953 Jul;203(1):153–159. [PubMed] [Google Scholar]
- Blattner F. R., Burland V., Plunkett G., 3rd, Sofia H. J., Daniels D. L. Analysis of the Escherichia coli genome. IV. DNA sequence of the region from 89.2 to 92.8 minutes. Nucleic Acids Res. 1993 Nov 25;21(23):5408–5417. doi: 10.1093/nar/21.23.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bulawa C. E., Raetz C. R. Isolation and characterization of Escherichia coli strains defective in CDP-diglyceride hydrolase. J Biol Chem. 1984 Sep 25;259(18):11257–11264. [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Chen Y. M., Zhu Y., Lin E. C. The organization of the fuc regulon specifying L-fucose dissimilation in Escherichia coli K12 as determined by gene cloning. Mol Gen Genet. 1987 Dec;210(2):331–337. doi: 10.1007/BF00325702. [DOI] [PubMed] [Google Scholar]
- Cole S. T., Eiglmeier K., Ahmed S., Honore N., Elmes L., Anderson W. F., Weiner J. H. Nucleotide sequence and gene-polypeptide relationships of the glpABC operon encoding the anaerobic sn-glycerol-3-phosphate dehydrogenase of Escherichia coli K-12. J Bacteriol. 1988 Jun;170(6):2448–2456. doi: 10.1128/jb.170.6.2448-2456.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiglmeier K., Boos W., Cole S. T. Nucleotide sequence and transcriptional startpoint of the glpT gene of Escherichia coli: extensive sequence homology of the glycerol-3-phosphate transport protein with components of the hexose-6-phosphate transport system. Mol Microbiol. 1987 Nov;1(3):251–258. doi: 10.1111/j.1365-2958.1987.tb01931.x. [DOI] [PubMed] [Google Scholar]
- Freedberg W. B., Kistler W. S., Lin E. C. Lethal synthesis of methylglyoxal by Escherichia coli during unregulated glycerol metabolism. J Bacteriol. 1971 Oct;108(1):137–144. doi: 10.1128/jb.108.1.137-144.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freedberg W. B., Lin E. C. Three kinds of controls affecting the expression of the glp regulon in Escherichia coli. J Bacteriol. 1973 Sep;115(3):816–823. doi: 10.1128/jb.115.3.816-823.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- GAREN A., LEVINTHAL C. A fine-structure genetic and chemical study of the enzyme alkaline phosphatase of E. coli. I. Purification and characterization of alkaline phosphatase. Biochim Biophys Acta. 1960 Mar 11;38:470–483. doi: 10.1016/0006-3002(60)91282-8. [DOI] [PubMed] [Google Scholar]
- HAYASHI S., LIN E. C. CAPTURE OF GLYCEROL BY CELLS OF ESCHERICHIA COLI. Biochim Biophys Acta. 1965 Mar 29;94:479–487. doi: 10.1016/0926-6585(65)90056-7. [DOI] [PubMed] [Google Scholar]
- Hazelbauer G. L. Maltose chemoreceptor of Escherichia coli. J Bacteriol. 1975 Apr;122(1):206–214. doi: 10.1128/jb.122.1.206-214.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin R. Z., Lin E. C. An inducible phosphoenolpyruvate: dihydroxyacetone phosphotransferase system in Escherichia coli. J Gen Microbiol. 1984 Jan;130(1):83–88. doi: 10.1099/00221287-130-1-83. [DOI] [PubMed] [Google Scholar]
- Kelley J. J., Dekker E. E. Identity of Escherichia coli D-1-amino-2-propanol:NAD+ oxidoreductase with E. coli glycerol dehydrogenase but not with Neisseria gonorrhoeae 1,2-propanediol:NAD+ oxidoreductase. J Bacteriol. 1985 Apr;162(1):170–175. doi: 10.1128/jb.162.1.170-175.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kistler W. S., Lin E. C. Anaerobic L- -glycerophosphate dehydrogenase of Escherichia coli: its genetic locus and its physiological role. J Bacteriol. 1971 Dec;108(3):1224–1234. doi: 10.1128/jb.108.3.1224-1234.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohara Y., Akiyama K., Isono K. The physical map of the whole E. coli chromosome: application of a new strategy for rapid analysis and sorting of a large genomic library. Cell. 1987 Jul 31;50(3):495–508. doi: 10.1016/0092-8674(87)90503-4. [DOI] [PubMed] [Google Scholar]
- Kuritzkes D. R., Zhang X. Y., Lin E. C. Use of phi(glp-lac) in studies of respiratory regulation of the Escherichia coli anaerobic sn-glycerol-3-phosphate dehydrogenase genes (glpAB). J Bacteriol. 1984 Feb;157(2):591–598. doi: 10.1128/jb.157.2.591-598.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Larson T. J., Schumacher G., Boos W. Identification of the glpT-encoded sn-glycerol-3-phosphate permease of Escherichia coli, an oligomeric integral membrane protein. J Bacteriol. 1982 Dec;152(3):1008–1021. doi: 10.1128/jb.152.3.1008-1021.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson T. J., Ye S. Z., Weissenborn D. L., Hoffmann H. J., Schweizer H. Purification and characterization of the repressor for the sn-glycerol 3-phosphate regulon of Escherichia coli K12. J Biol Chem. 1987 Nov 25;262(33):15869–15874. [PubMed] [Google Scholar]
- Lin E. C. Glycerol dissimilation and its regulation in bacteria. Annu Rev Microbiol. 1976;30:535–578. doi: 10.1146/annurev.mi.30.100176.002535. [DOI] [PubMed] [Google Scholar]
- Meagher R. B., Tait R. C., Betlach M., Boyer H. W. Protein expression in E. coli minicells by recombinant plasmids. Cell. 1977 Mar;10(3):521–536. doi: 10.1016/0092-8674(77)90039-3. [DOI] [PubMed] [Google Scholar]
- Sanno Y., Wilson T. H., Lin E. C. Control of permeation to glycerol in cells of Escherichia coli. Biochem Biophys Res Commun. 1968 Jul 26;32(2):344–349. doi: 10.1016/0006-291x(68)90392-6. [DOI] [PubMed] [Google Scholar]
- Schweizer H., Boos W., Larson T. J. Repressor for the sn-glycerol-3-phosphate regulon of Escherichia coli K-12: cloning of the glpR gene and identification of its product. J Bacteriol. 1985 Feb;161(2):563–566. doi: 10.1128/jb.161.2.563-566.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprenger G. A., Hammer B. A., Johnson E. A., Lin E. C. Anaerobic growth of Escherichia coli on glycerol by importing genes of the dha regulon from Klebsiella pneumoniae. J Gen Microbiol. 1989 May;135(5):1255–1262. doi: 10.1099/00221287-135-5-1255. [DOI] [PubMed] [Google Scholar]
- St Martin E. J., Freedberg W. B., Lin E. C. Kinase replacement by a dehydrogenase for Escherichia coli glycerol utilization. J Bacteriol. 1977 Sep;131(3):1026–1028. doi: 10.1128/jb.131.3.1026-1028.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweet G., Gandor C., Voegele R., Wittekindt N., Beuerle J., Truniger V., Lin E. C., Boos W. Glycerol facilitator of Escherichia coli: cloning of glpF and identification of the glpF product. J Bacteriol. 1990 Jan;172(1):424–430. doi: 10.1128/jb.172.1.424-430.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeshita S., Sato M., Toba M., Masahashi W., Hashimoto-Gotoh T. High-copy-number and low-copy-number plasmid vectors for lacZ alpha-complementation and chloramphenicol- or kanamycin-resistance selection. Gene. 1987;61(1):63–74. doi: 10.1016/0378-1119(87)90365-9. [DOI] [PubMed] [Google Scholar]
- Tang C. T., Ruch F. E., Jr, Lin C. C. Purification and properties of a nicotinamide adenine dinucleotide-linked dehydrogenase that serves an Escherichia coli mutant for glycerol catabolism. J Bacteriol. 1979 Oct;140(1):182–187. doi: 10.1128/jb.140.1.182-187.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang J. C., Forage R. G., Lin E. C. Immunochemical properties of NAD+-linked glycerol dehydrogenases from Escherichia coli and Klebsiella pneumoniae. J Bacteriol. 1982 Dec;152(3):1169–1174. doi: 10.1128/jb.152.3.1169-1174.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truniger V., Boos W., Sweet G. Molecular analysis of the glpFKX regions of Escherichia coli and Shigella flexneri. J Bacteriol. 1992 Nov;174(21):6981–6991. doi: 10.1128/jb.174.21.6981-6991.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voegele R. T., Sweet G. D., Boos W. Glycerol kinase of Escherichia coli is activated by interaction with the glycerol facilitator. J Bacteriol. 1993 Feb;175(4):1087–1094. doi: 10.1128/jb.175.4.1087-1094.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]