Abstract
We report here a method for the in vivo dissection of the regulatory region of a gene in the Drosophila genome. Our system includes (i) the reporter genes lacZ and white to detect transcriptional enhancer and silencer activities in a target gene, (ii) an efficient way to induce integration of gypsy elements in the genome, and (iii) unidirectional blocking of regulatory activities by the gypsy element, which is dependent on the su(Hw) protein. The optomotor-blind (omb) gene was analyzed. In the ombP1 line, a P[lacW] construct is inserted about 1.4 kb upstream of the omb transcription start site. The lacZ reporter gene within P[lacW] exhibits the same expression pattern as omb. The white reporter gene is expressed in a “bipolar” pattern. We induced high frequency gypsy mobilization in ombP1 and identified two lines (D11 and D13–1) with altered eye pigmentation pattern, which is dependent on su(Hw) activity. A gypsy element was found inserted in the first intron of omb in D13–1 and in P[lacW] in D11. These results indicate that it is the blocking of regulatory activities by gypsy that caused the changes in the white reporter gene expression. The effect of these gypsy insertions on the expression patterns allowed us to predict several aspects of the organization of the regulatory elements in the omb locus.
Keywords: gypsy, enhancer, silencer, transcriptional regulation
The usual approach to analyzing the cis-acting regulatory region of a gene is to clone fragments of the region into a tester vector with a reporter gene and analyze its effect in cultured cells or transgenic organisms. Although fruitful for testing the sufficiency of an enhancer element, this approach takes the fragment out of its normal chromosomal context. It is also not suitable for a detailed analysis of a large regulatory region, which is common to many developmentally important genes. We report here a new approach for the in vivo analysis of the cis-acting regulatory region within its normal chromosomal context.
The approach was used to analyze the regulatory region of the large and genetically complex optomotor-blind (omb) gene in Drosophila melanogaster. The omb gene spans at least 120 kb, with a 70-kb transcription unit and a 6-kb mature transcript (1). It encodes a DNA binding protein (2). omb is expressed in specific patterns in embryo, larval imaginal discs, and brain (3). Mutations affecting the development of adult wing, optic lobe, and abdominal tergite pigmentation have been mapped to regions upstream, downstream, and within the transcription unit (4–6), suggesting that there are multiple regulatory elements responsible for omb expression in different tissues. An enhancer trap line ombP1 was identified to have a P[lacW] (7) inserted in omb. P[lacW] carries two genes, mini-white (w+m) and lacZ, that serve as reporters of local enhancer/silencer activities. Our preliminary results suggested that the w+m expression pattern in ombP1 is sensitive to changes in local chromosomal environment (Y.H.S., unpublished results), and thus is suited for our analysis.
Many spontaneous mutations in D. melanogaster were found to be due to insertion of the gypsy transposable element. It need not to insert within a transcription unit for its mutational effect. The gypsy element contains a binding region for the su(Hw) protein (8–10). Binding to the su(Hw) protein (SUHW) blocks the activity of transcriptional enhancers and thus interferes with the expression of the gene adjacent to gypsy insertion. Only the enhancers located distal (relative to the promoter) to the su(Hw) binding region are blocked (11, 12). This unidirectional blocking effect also works with the repressive activity of heterochromatin, telomeric chromatin, chromosomal position-effect, and Polycomb response elements (13, 14, ‖), but not with a zen ventral silencer (16).
The gypsy elements were recently shown to be infectious retrovirus (17, 18). Transposition of gypsy is controlled by the flamenco gene (flam), located on the X chromosome (19). Fly strains carrying a flam permissive allele and functional gypsy provirus can produce infectious retroviral particles in the somatic follicle cells in the ovaries (20). These particles then infect the oocytes, and gypsy insertion is seen in the progeny of flam/flam homozygous mutant females.
In this report, we took advantage of these new findings to generate random gypsy insertions in the ombP1 line. If gypsy inserts in the omb locus and blocks the activity of certain regulatory elements, the expression pattern of the mini-white (w+m) and lacZ reporter genes may be affected. Two such lines were isolated and characterized. The insertion sites of gypsy within the omb locus were mapped, and the change in eye color pattern was shown to be dependent on su(Hw) activity. These results allowed the mapping of discrete regulatory elements in the omb locus.
MATERIALS AND METHODS
Drosophila Stocks.
w ombP1 was previously described (21). cm ct6 sn4; su(Hw)f TM6/su(Hw)2 sbd, FM3/y v f mal flam1, and C(1)Dx, y f/y w v f mal flam1 were kindly provided by Alain Bucheton (Centre de Genetique Moleculaire, Centre National de la Recherche Scientifique, Gif-Sur-Yvette, France). Mutations are described in Lindsley and Zimm (22).
Gypsy Mobilization.
Gypsy transposition was induced by using the flamenco (flam) line, which is permissive to the formation of infectious gypsy viral particles (19, 20). Homozygous y w v f mal flam1 females were mated to y w ombP1 males. The F1 y w ombP1/y w v f mal flam1 females, now carrying germ-line gypsy insertions, were mated to FM0/Y males. About 5850 F2 y w ombP1/FM0 females and y w ombP1/Y males were screened for changes in the bipolar eye color pattern. Screening in the females would enable the isolation of omb lethal mutations. This is possible because the bipolar eye color pattern is a dominant phenotype.
Molecular Analysis.
A 0.9-kb fragment (pX35) was cloned by plasmid rescue from EcoRI-digested ombP1 genomic DNA. A genomic λ clone (λX35.6) encompassing the site of P[lacW] insertion in ombP1 was isolated from a Canton-S library in λCh4A (23). The four EcoRI fragments from this clone were subcloned and used as probe in genome blot analysis. The E1.8kb probe detected size changes in D13–1; the 5.6-kb EcoRI fragment was changed to 4.1 kb, and the 5.0-kb HindIII fragment was changed to 7.1 kb. These changes can be mapped to within the 0.7-kb PstI/HindIII fragment. In D11, the E1.8kb probe detected no change in EcoRI and HindIII fragments, but there was a change of the 12.5-kb PstI fragment to 9.4 kb. Thus, the change in D11 can be mapped to within the 10.5-kb fragment from the PstI site in the P[lacW] polylinker to the first HindIII site downstream of P[lacW] insert.
PCR (Expand Long Template PCR, Boehringer Mannheim) was used to amplify DNA sequences between P[lacW] and the gypsy element. Oligonucleotide primers X1–X6 extend outward from the long terminal repeat (LTR) region of gypsy. The position (5′ to 3′) of X1–X6 in the published gypsy sequence (24) are: X1, 224–201; X2, 282–264; X3, 7141–7160; X4, 7169–7186; X5, 108–90; X6, 7350–7370. Four primers from the terminal region of P element were used. P31 extends outward: 5′-CGACGGGACCACCTTATGTTATTTCATCATG-3′. XP37 is P31 with an added 5′ XhoI site. P6/31 extends inward: 5′-TGAAATAACATAAGGTGGTCCCGTCG-3′. P31/6 is the reverse of P6/31. PCR was carried out with a Perkin–Elmer PE9600 using the following program: 92°C, 2 min; 10 cycles of 92°C, 10 sec; 55°C, 30 sec; 68°C, 15 min; 15 cycles of 92°C, 10 sec; 55°C, 30 sec; 68°C, 15 min plus 20-sec increments per cycle, and a final extension at 68°C for 7 min.
In D13–1, nested PCR using primers extending outward from the gypsy LTR and primers extending outward from the P-element terminus (X3–P37 and X4–XP37) generated a 5.7-kb fragment. Hybridization with the E1.7kb and E1.8kb probes were positive, confirming that this fragment is derived from the omb locus. It was cloned into the pGEM-T TA cloning vector (Promega). Terminal sequences confirmed that one end corresponds to the P[lacW]-omb junction, the other end is from the gypsy LTR and extends into probably the unsequenced omb first intron. Terminal sequencing of two subclones (3.4 kb EcoRI/XhoI and 4.4 kb HindIII/XhoI fragments subcloned into pBluescript KS) also identified omb sequences. Based on these data, the gypsy insertion site in D13–1 is about 4.2 kb downstream from the first exon and within the large first intron of omb.
The above PCR strategy failed to amplify any fragment in D11, suggesting that the gypsy insertion may be within P[lacW]. A primer (P6/31) extending inward from the P terminus was used in nested PCR with gypsy LTR primers (X2/X1 and X3/X4), and generated a 6.5-kb and a 4-kb fragment, respectively. Direct PCR sequencing (according to Risinger et al. (25), using X5 and X6 primers, respectively) showed that the gypsy is inserted in the lacZ gene (between bases 3358 and 3359, CATGTATA/CCCCGT, in the pP[lacW] coordinate in ref. 26) and created a 4-bp (TATA) duplication of target site, consistent with the known length of the target site duplication of gypsy (27).
X-Gal (5-bromo-4-chloro-3-indolyl β-d-galactoside) Staining.
Embryos were stained according to Hiromi et al. (28), but without devitellination. Staining of imaginal discs was according to Sun et al. (21).
RESULTS
The ombP1 Line.
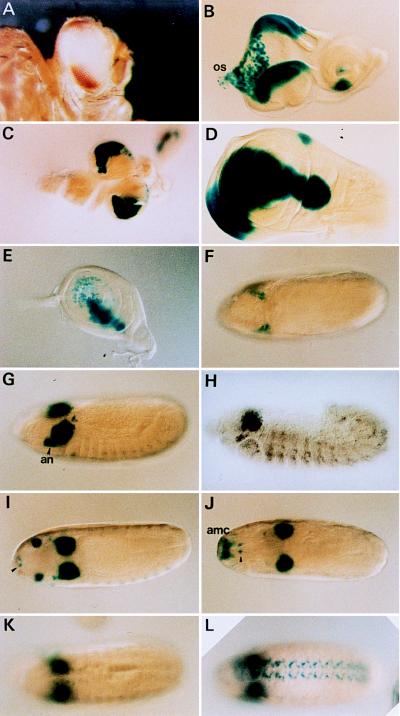
The ombP1 line is a P[lacW] transposant line (21). In situ hybridization to polytene chromosomes mapped the P[lacW] to 4C3–6 (21). P[lacW] carries two genes that serve as the reporter of enhancer/silencer activities: mini-white (w+m) is responsive to silencer activity and lacZ is responsive to enhancer activity. In ombP1 lacZ expression apparently reflects the omb expression pattern in embryos and in imaginal discs (as determined by in situ hybridization in ref. 3 and anti-OMB (optomotor-blind) antibody staining in ref. 29), with only minor differences (Fig. 1). w+m expression is suppressed in the central part of the eye, leaving only the dorsal and ventral poles pigmented (Fig. 1A). This “bipolar” eye color pattern provides an easily scorable phenotype, which reflects the spatial expression pattern of the w+m reporter gene.
Figure 1.
Expression patterns of reporter genes in ombP1 (A–G and I–K) ombP1, (H) wild type, (L) D13–1. (A) The bipolar w+m expression pattern in ombP1. (B–G and H–K) lacZ expression pattern as stained by X-Gal. The pattern is similar to omb expression (3, 29), except when noted below. (B) In eye–antennal disc expression is in the dorsal and ventral poles of the eye disc, primarily in the peripodial membrane destined to become part of the head capsule (30). Expression is also seen in scattered cells in the proximal region of the disc and the optic stalk (os), and in a ventral sector in the antennal disc. (C) Expression in the central nervous system is in the optic lobe, plus a few cells in the ventral ganglia. (D) Expression in the wing disc is in a broad domain straddling the anterior–posterior compartmental boundary. This expression has recently been shown to be under dpp and wg regulation (29, 31, 32). Weak omb expression in the notal region is not detected in lacZ expression (compare with figure 2C in ref. 29). (E) In the leg disc, expression is also in a stripe along the anterior–posterior compartmental boundary, but only in the dorsal region. (F) Dorsal view of a stage 8 embryo. Expression is in two lateral clusters of cells anterior to the cephalic furrow. (G) Slightly oblique lateral view of a stage 12 embryo. Expression is strong in the optic lobe anlage and antennal segment (an). The lateral segmentally repeating pattern is weaker than omb (compare with H). (H) Expression detected by in situ hybridization of omb in a wild-type embryo of similar stage as in G. The antennal expression is weaker. (I) Dorsal view at about stage 13 (before head involution); the antennal expression had moved dorsally and anteriorly. Expression also occurred in two ventral anterior spots, probably the hypopharyngeal sensory organ (ho; arrowhead). (J) After head involution, the antennal expression spot had moved to a position corresponding to the antennomaxillary complex (amc) (dorsal view). (K) X-Gal staining in the ventral nerve cord is weaker compared with omb in situ hybridization in wild type (compare with figure 2H in ref. 3). (L) The ventral nerve cord expression is stronger in D13–1. (K and L) Dorsal view.
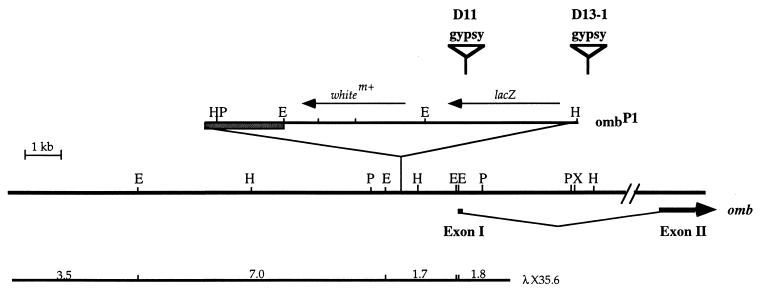
The genomic fragment flanking the P[lacW] insertion site was cloned by plasmid rescue. DNA sequence obtained from the two ends of the fragment matched with sequence of the omb locus, placing the site of insertion 1373 bp upstream of the omb cDNA 5′ end (Fig. 2). An 8-bp duplication (CCACAGTC) of the target site was created.
Figure 2.
Site of P[lacW] insertion in ombP1, and the site of gypsy insertion in D13–1 and D11. The site of the P[lacW] insertion in ombP1 is indicated on the genomic map of omb. The cross-hatched box indicates the plasmid portion of P[lacW]. Four EcoRI fragments from a genomic λ clone (λX35.6) were subcloned and used as probe in genome blot analysis. The sites of gypsy insertion in D11 and D13–1 are indicated. The gypsy element is not drawn to scale.
Isolation of Lines with Altered Eye Pigmentation Pattern.
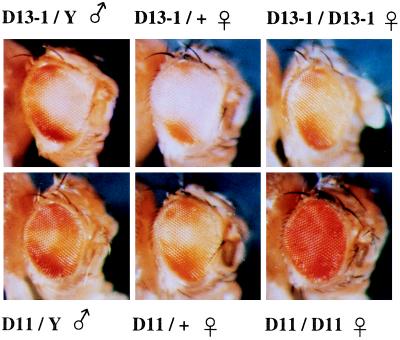
Gypsy transposition was induced using the permissive flamenco (flam) line and screened for insertion into the y w ombP1 chromosome and changes in the bipolar eye color pattern. Two independent lines (D11 and D13–1) were isolated that have altered eye color pattern (Fig. 3), which is dependent on su(Hw) activity (see the later). In D11 males, the polar pigmented domains have expanded and the interpolar region is lightly pigmented. In D13–1 males, the posterior rim between the two poles is pigmented. The cause of the eye color pattern change was determined by segregation analysis to be on the X chromosome. In repeated outcrosses of heterozygous D11 and D13–1 females to w males, no fly with the original bipolar pattern was observed. Recombination with cm ct sn gave frequencies consistent with the map location of omb (data not shown), also suggestive of a cis-acting effect in the omb locus.
Figure 3.
Pattern of w+m reporter expression in the gypsy insertion lines. (Upper) D13–1. (Lower) D11.
Pattern Change Is Dependent on su(Hw) Function.
If the phenotype change is due to a gypsy insertion into the omb locus and the blocking of regulatory activity, then it may be reversed by su(Hw) mutation. The mutants were crossed to a su(Hw) mutant. About one-quarter of the F2 non-w+ male progeny have the original bipolar pattern (not shown, compare with Fig. 1A), suggesting that the phenotype was suppressed in the su(Hw) mutant background. su(Hw) mutation had no effect on ombP1, demonstrating that the effect is not on P[lacW].
To confirm the su(Hw) effect, D11 and D13–1 were recombined onto a ct6 chromosome, and the experiment with su(Hw) repeated. ct6 is a mutation due to gypsy insertion into the cut locus, and its phenotypic reversion serves as an indicator of the su(Hw)f/su(Hw)2 genotype. The reversion of D13–1 and D11 to the original bipolar pattern is always accompanied by reversion of the ct phenotype. This unambiguously shows that the phenotypic change is dependent on the wild-type su(Hw) function, most likely acting through a gypsy insertion in the omb locus.
Mapping of Gypsy Insertions in the omb Locus.
A genomic λ clone (λX35.6) encompassing the site of P[lacW] insertion in ombP1 was isolated (Fig. 2). The EcoRI fragments from this clone were subcloned and used as probes in genome blot analysis, comparing D13–1, D11, and ombP1 (data not shown). Fragment size changes were detected and mapped in D13–1 and D11 (Fig. 2), suggesting the insertion of gypsy. Long range PCR was used to clone the fragments between gypsy and P[lacW] from D13–1 and D11. Terminal sequencing of the PCR fragments mapped the gypsy insertion site in D13–1 at about 4.2 kb downstream from the first exon and within the large first intron of omb (Fig. 2). In D11, the gypsy is inserted in the lacZ gene in the P[lacW] (Fig. 2).
Expression and Phenotype of D13–1 and D11.
D13–1 is hemizygous and homozygous viable and showed no apparent morphological abnormality. Weak wing vein delta formation occurs when in combination with a strong hypomorphic mutant ombN76 (data not shown), indicating that it is a weak hypomorphic omb mutation. X-Gal staining of D13–1 showed that the expression pattern and intensity of the lacZ reporter gene in embryos and in imaginal discs are indistinguishable from ombP1, except that the embryonic ventral nerve cord expression in D13–1 is enhanced (Fig. 1L).
D11 is hemizygous viable and exhibits only a weak wing vein phenotype. D11 showed no X-Gal staining in embryos or in imaginal discs, consistent with the finding that the lacZ gene is interrupted by the gypsy. However, gypsy is inserted near the 3′ end of lacZ, so it should leave most of the lacZ transcript detectable. In situ hybridization in ombP1 embryo detected lacZ expression in the optic lobe and the antennomaxillary organ anlage (not shown), although much weaker than X-Gal staining. The weaker expression in the ventral nerve cord and hypopharyngeal sensory organ were not detectable. D11 embryo showed a similar lacZ expression pattern, except that the signal is slightly weaker (not shown).
In both D11 and D13–1, homozygous females have an enhanced eye pigmentation compared with hemizygous males and heterozygous females (Fig. 3), suggesting a pairing effect on the w+m reporter expression, an effect which was not observed in ombP1. D13–1 females, but not males, show an extended pigmentation of the abdominal tergites, comparable to the phenotype of Qd-type omb alleles (33). Similar transvection effect of Qd phenotype has been reported (33).
DISCUSSION
Our results pinpointed the insertion of a gypsy element in the first intron of the omb gene in D13–1, and in the middle of lacZ in P[lacW] in D11. These insertions caused the bipolar eye pattern in ombP1 to change. The effect is dependent on su(Hw) function. The simplest explanation is that the binding of SUHW to its target site in the gypsy element blocked the activity of some silencer located distal (relative to the promoter) of the gypsy insertion sites. In contrast to a previous report that the su(Hw) binding region is unable to block the 600-bp zen ventral silencer element (16), our results provide evidence that at least some silencer–promoter interaction can be blocked.
D13–1 is a weak hypomorphic mutation. The gypsy insertion in the intron probably leads to a reduced level of the correctly spliced transcript, thus causing the weak wing phenotype but no lethality. In the omb hypomorph bifid (ombbi), the wing phenotype is also caused by an insertion into the first omb intron (33). The lacZ expression patterns in D13–1 and ombP1 embryos, imaginal discs, and brain are indistinguishable, except that the embryonic ventral nerve cord expression in D13–1 is enhanced. If gypsy serves as a general and efficient enhancer block and has strict polarity, we can conclude that in the region downstream of the D13–1 gypsy insertion site there is (i) no strong enhancer for embryo and imaginal disc expression, (ii) a silencer acting in the posterior rim of the eye disc, (iii) a silencer acting in the embryonic ventral nerve cord, and (iv) an element suppressing the pairing effect.
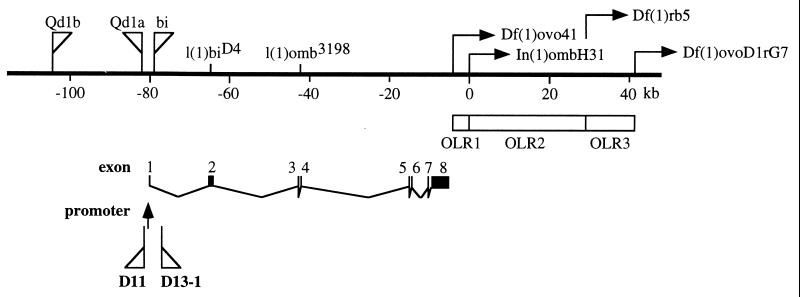
Chromosomal rearrangements that remove the 3′ end of the omb locus cause specific anatomical defects in the optic lobe. Three optic lobe regulatory regions (OLRs) were defined (ref. 6; Fig. 4). In In(1)ombH31, omb expression in the optic lobe (OL) anlage is reduced in late embryos and during larval development (3). We did not observe, however, obvious changes in the OL expression pattern in D13–1. These discrepancies can be interpreted in several ways. First, there may be an additional promoter located downstream of the D13–1 insertion so that D13–1 would not block larval regulation from the downstream OLR elements. Second, there may be a set of enhancer (OLRs) and silencers acting in late embryonic OL. In(1)ombH31 removed the enhancer, leading to reduced expression and defective OL. D13–1 blocked both, and so caused no change in expression level. Third, the blocking by gypsy may not be as efficient as expected.
Figure 4.
Map of the omb locus. The genetic map of omb is redrawn from ref. 33. l(1)biD4 and l(1)omb3198 are two point mutations causing premature termination of the omb protein (34). Qd1a, Qd1b, and bi indicate insertions found in the Qd and bi mutant chromosome, respectively. OLRs indicate optic lobe regulatory regions defined by the various chromosomal breakpoints. The sites of gypsy insertion in D11 and D13–1 are indicated relative to the promoter.
The gypsy in D11 is inserted in the P[lacW], which inserts 1373 bp upstream of the 5′ end of the omb cDNA. D11 is hemizygous viable and exhibits only a weak wing vein phenotype, indicating that the gypsy insertion did not block any essential regulatory element. In addition, the gypsy insertion apparently blocked a silencer acting in the eye disc and an element repressing the pairing effect. The gypsy insertion site is upstream of the transcription start site (G.O.P., unpublished results). This suggests that the 5.6-kb region between the two gypsy insertion sites (D11 and D13–1) contained most of the regulatory elements. Alternatively, as proposed above, there is an additional downstream promoter so that D11 and D13–1 are both upstream of the promoter, and thus have similar effects.
The approach presented in this study allows the efficient analysis of a large and complex regulatory region. The regulatory elements are analyzed in their normal chromosomal location, within a normal context of other regulatory elements. The effect on expression is detected in an organism, rather than in tissue culture cells or in vitro. The effect on expression (of the lacZ reporter) can be monitored in different developmental stages and in different tissues. Roseman et al. (14) has reported the use of a similar approach by examining the phenotypic effect of su(Hw) mediated blocking.
The w+m reporter gene provides an easily scorable dominant phenotype (eye color). This made possible the screening of a large number of flies heterozygous for the potential mutation. It is likely that a mutation affecting the regulatory region of an important gene may lead to lethality. The ability to screen in the heterozygous condition will allow the isolation of recessive lethal mutations.
While we demonstrated that the gypsy element can be used as a tool for the in vivo analysis of regulatory region and for mutagenesis, gypsy insertions cannot be easily manipulated further. A P-element construct carrying the su(Hw) binding region and the lacZ and w+m reporter genes might be used instead of gypsy (see also ref. 14). It can serve to block regulatory activities on one side and detect regulatory activities on the other side of its insertion site in a gene. Once inserted into a locus, local jumps (35, 36) can be induced at high frequency and changes in lacZ expression can be screened directly. A series of such insertions in a locus would be equivalent to a series of progressive terminal deletions, thereby providing a high-resolution map of the regulatory region. Dependence on SUHW protein provides an easy way to check whether any effect on gene expression is indeed due to the blockage. Development of such a system is in progress.
Finally, other boundary elements similar in function to the su(Hw) binding region have been identified, e.g., the 5′ constitutive-hypersensitive site from the chicken β-globin domain (37), the scs, scs′, Mcp, and Fab-7 elements in Drosophila (15, 38–41). Taking a strategy similar to the one described here, these boundary elements might be used for mutagenesis and unidirectional blocking of regulatory activity in organisms other than Drosophila. What is required is a reporter gene sensitive to regulatory activities, and a construct carrying the blocking element and capable of random integration into the genome (e.g., a retroviral construct). Targeted insertion via homologous recombination in embryonic stem cells may be used to study the regulatory region of a specific gene.
Acknowledgments
We would like to thank Dr. Alain Bucheton for providing the flam and su(Hw) fly stocks, Chiou-Yang Tang and Susan Hshieh for maintaining fly stocks, and Burkhard Poeck for providing the omb in situ hybridization data, and Stefan Grimm for help in the preparation of the figures. We thank an anonymous reviewer for suggesting lacZ in situ hybridization of D11. This study was supported by grants to Y.H.S. from the Academia Sinica (5202401023–4) and from the National Science Council (NSC83–2311-B001–110) of the Republic of China, by a grant to S.-F.T. (NSC84–2331-B010–017) from the National Science Council of the Republic of China, and by a grant to G.O.P. from Deutsche Forschungsgemeinschaft (Pf 163/6–3).
ABBREVIATIONS
- LTR
long terminal repeat
- X-Gal
5-bromo-4-chloro-3-indolyl β-d-galactoside
- OLR
optic lobe regulatory region
- OL
optic lobe
Footnotes
Pirrotta, V., Poux, S., Kostic, C., Sigrist, C., Tatout, C. & Horard, B., 37th Drosophila Research Conference, April 27–May 1, 1996, Chicago, p. 45.
References
- 1.Pflugfelder G O, Schwarz H, Roth H, Poeck B, Sigl A, Kerscher S, Jonschker B, Pak W L, Heisenberg M. Genetics. 1990;126:91–104. doi: 10.1093/genetics/126.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pflugfelder G O, Roth H, Poeck B. Biochem Biophys Res Commun. 1992;186:918–925. doi: 10.1016/0006-291x(92)90833-7. [DOI] [PubMed] [Google Scholar]
- 3.Poeck B, Hofbauer A, Pflugfelder G O. Development (Cambridge, UK) 1993;117:1017–1029. doi: 10.1242/dev.117.3.1017. [DOI] [PubMed] [Google Scholar]
- 4.Heisenberg M, Wonneberger R, Wolf R. J Comp Physiol. 1978;124:287–296. [Google Scholar]
- 5.Pflugfelder G O, Roth H, Poeck B, Kerscher S, Schwarz H, Jonschker B, Heisenberg M. Proc Natl Acad Sci USA. 1992;89:1199–1203. doi: 10.1073/pnas.89.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brunner A, Wolf R, Pflugfelder G O, Poeck B, Heisenberg M. J Neurogenet. 1992;8:43–55. doi: 10.3109/01677069209167271. [DOI] [PubMed] [Google Scholar]
- 7.Bier E, Vaessin H, Shepherd S, Lee K, McCall K, Barbel S, Ackerman L, Carretto R, Uemura T, Grell E, Jan L Y, Jan Y N. Genes Dev. 1989;3:1273–1287. doi: 10.1101/gad.3.9.1273. [DOI] [PubMed] [Google Scholar]
- 8.Geyer P K, Green M M, Corces V G. Proc Natl Acad Sci USA. 1988;85:8593–8597. doi: 10.1073/pnas.85.22.8593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Peifer M, Bender W. Proc Natl Acad Sci USA. 1988;85:9650–9654. doi: 10.1073/pnas.85.24.9650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Spana C, Harrison D A, Corces V G. Genes Dev. 1988;2:1414–1423. doi: 10.1101/gad.2.11.1414. [DOI] [PubMed] [Google Scholar]
- 11.Geyer P K, Corces V G. Genes Dev. 1992;6:1865–1873. doi: 10.1101/gad.6.10.1865. [DOI] [PubMed] [Google Scholar]
- 12.Dorsett D. Genetics. 1993;134:1135–1144. doi: 10.1093/genetics/134.4.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Roseman R R, Pirrotta V, Geyer P. EMBO J. 1993;12:435–442. doi: 10.1002/j.1460-2075.1993.tb05675.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Roseman R R, Johnson E A, Rodesch C K, Bjerke M, Nagoshi R N, Geyer P K. Genetics. 1995;141:1061–1074. doi: 10.1093/genetics/141.3.1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao K, Hart C, Laemmli U. Cell. 1996;81:879–889. doi: 10.1016/0092-8674(95)90008-x. [DOI] [PubMed] [Google Scholar]
- 16.Cai H, Levine M. Nature (London) 1995;376:533–536. doi: 10.1038/376533a0. [DOI] [PubMed] [Google Scholar]
- 17.Kim A, Terzian C, Santamaria P, Pelisson A, Prud’homme N, Bucheton A. Proc Natl Acad Sci USA. 1994;91:1285–1289. doi: 10.1073/pnas.91.4.1285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Song S U, Gerasimova T, Kurkulos M, Boeke J D, Corces V G. Genes Dev. 1994;8:2046–2057. doi: 10.1101/gad.8.17.2046. [DOI] [PubMed] [Google Scholar]
- 19.Prud’homme N, Gans M, Masson M, Terzian C, Bucheton A. Genetics. 1995;139:697–711. doi: 10.1093/genetics/139.2.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pelisson A, Song S U, Prud’homme N, Smith P A, Bucheton A, Corces V G. EMBO J. 1994;13:4401–4411. doi: 10.1002/j.1460-2075.1994.tb06760.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sun Y H, Tsai C-J, Green M M, Chao J-L, Yu C-T, Jaw T J, Yeh J-Y, Bolshakov V N. Genetics. 1995;141:1075–1086. doi: 10.1093/genetics/141.3.1075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindsley D I, Zimm G G. The Genome of Drosophila melanogaster. New York: Academic; 1992. [Google Scholar]
- 23.Maniatis T, Hardison R C, Lacy E, Lauer J, O’Connell C, Quon D. Cell. 1978;15:687–701. doi: 10.1016/0092-8674(78)90036-3. [DOI] [PubMed] [Google Scholar]
- 24.Marlor R L, Parkhurst S M, Corces V G. Mol Cell Biol. 1986;6:1129–1134. doi: 10.1128/mcb.6.4.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Risinger J I, Berchuck A, Kohler M F, Boyd J. Nat Genet. 1994;7:98–102. doi: 10.1038/ng0594-98. [DOI] [PubMed] [Google Scholar]
- 26.FlyBase. Nucleic Acids Res. 1996;24:53–56. doi: 10.1093/nar/24.1.53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freund R, Meselson M. Proc Natl Acad Sci USA. 1984;81:4462–4464. doi: 10.1073/pnas.81.14.4462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiromi Y, Kuroiwa A, Gehring W. Cell. 1985;43:603–613. doi: 10.1016/0092-8674(85)90232-6. [DOI] [PubMed] [Google Scholar]
- 29.Grimm S, Pflugfelder G O. Science. 1996;271:1601–1604. doi: 10.1126/science.271.5255.1601. [DOI] [PubMed] [Google Scholar]
- 30.Haynie J L, Bryant P J. Wilhelm Roux’s Arch Dev Biol. 1986;183:85–100. doi: 10.1007/BF00848779. [DOI] [PubMed] [Google Scholar]
- 31.Nellen D, Burke R, Struhl G, Basler K. Cell. 1996;85:357–368. doi: 10.1016/s0092-8674(00)81114-9. [DOI] [PubMed] [Google Scholar]
- 32.Lecuit T, Brook W J, Ng M, Calleja M, Sun H, Cohen S M. Nature (London) 1996;381:387–393. doi: 10.1038/381387a0. [DOI] [PubMed] [Google Scholar]
- 33.Pflugfelder G O, Heisenberg M. Comp Biochem Physiol. 1995;110:185–202. doi: 10.1016/0300-9629(94)00159-q. [DOI] [PubMed] [Google Scholar]
- 34.Poeck B, Balles J, Pflugfelder G O. Mol Gen Genet. 1993;238:325–332. doi: 10.1007/BF00291990. [DOI] [PubMed] [Google Scholar]
- 35.Tower J, Karpen G H, Craig N, Spradling A C. Genetics. 1993;133:347–359. doi: 10.1093/genetics/133.2.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang P, Spradling A C. Genetics. 1993;133:361–373. doi: 10.1093/genetics/133.2.361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chung J H, Whiteley M, Felsenfeld G. Cell. 1993;74:505–514. doi: 10.1016/0092-8674(93)80052-g. [DOI] [PubMed] [Google Scholar]
- 38.Udvardy A, Maine E, Schedl P. J Mol Biol. 1985;185:341–358. doi: 10.1016/0022-2836(85)90408-5. [DOI] [PubMed] [Google Scholar]
- 39.Kellum R, Schedl P. Mol Cell Biol. 1992;12:2424–2431. doi: 10.1128/mcb.12.5.2424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Galloni M, Gyurkovics H, Schedl P, Karch F. EMBO J. 1993;12:1087–1097. doi: 10.1002/j.1460-2075.1993.tb05750.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Karch F, Galloni M, Sipos L, Gausz J, Gyurkovics H, Schedl P. Nucleic Acids Res. 1994;22:3138–3146. doi: 10.1093/nar/22.15.3138. [DOI] [PMC free article] [PubMed] [Google Scholar]