Abstract
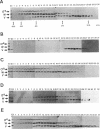
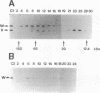
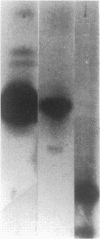
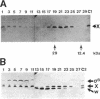
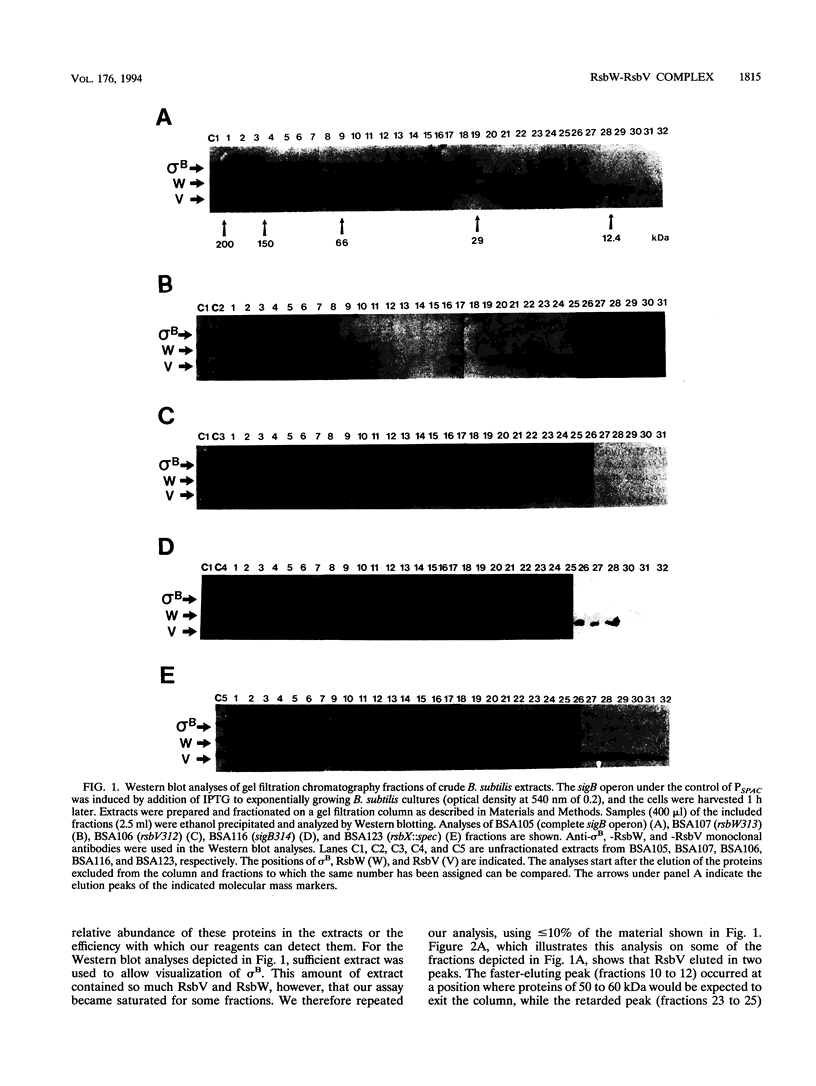
The activity of sigma B, a secondary sigma factor of Bacillus subtilis, is primarily controlled by an anti-sigma factor protein (RsbW) that binds to sigma B and blocks its ability to form an RNA polymerase holoenzyme (E-sigma B). Inhibition of sigma B by RsbW is counteracted by RsbV, a protein that is essential for the activation of sigma B-dependent transcription. When crude B. subtilis extracts were fractionated by gel filtration chromatography or electrophoresis through nondenaturing polyacrylamide gels, a complex composed of RsbW and RsbV that is distinct from the previously observed RsbW-sigma B complex was detected. In analogous experiments, RsbX, an additional regulator of sigma B-dependent transcription that is thought to act independently of RsbV-RsbW, was not found to associate with any of the other sigB operon products. Two forms of RsbV were visualized when crude cell extracts of B. subtilis were subjected to isoelectric focusing (IEF), with the more negatively charged RsbV species absent from extracts prepared from RsbW- strains. In vitro, RsbV became phosphorylated when incubated with ATP and RsbW but not with ATP alone. The phosphorylated RsbV species comigrated during IEF with the RsbW-dependent form of RsbV found in crude cell extracts. These results suggest that the modified RsbV, present in crude cell extracts, is phosphorylated. When gel filtration fractions containing RsbV-RsbW complexes or RsbV alone were subjected to IEF, only the unmodified form of RsbV was found associated with RsbW. The presumed phosphorylated variant of RsbV was present only in fractions that did not contain RsbW. The data support a model whereby RsbV binds directly to RsbW and blocks its ability to form the RsbW-sigma B complex. This activity of RsbV appears to be inhibited by RsbW-dependent phosphorylation.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benson A. K., Haldenwang W. G. Bacillus subtilis sigma B is regulated by a binding protein (RsbW) that blocks its association with core RNA polymerase. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2330–2334. doi: 10.1073/pnas.90.6.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson A. K., Haldenwang W. G. Characterization of a regulatory network that controls sigma B expression in Bacillus subtilis. J Bacteriol. 1992 Feb;174(3):749–757. doi: 10.1128/jb.174.3.749-757.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson A. K., Haldenwang W. G. Regulation of sigma B levels and activity in Bacillus subtilis. J Bacteriol. 1993 Apr;175(8):2347–2356. doi: 10.1128/jb.175.8.2347-2356.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson A. K., Haldenwang W. G. The sigma B-dependent promoter of the Bacillus subtilis sigB operon is induced by heat shock. J Bacteriol. 1993 Apr;175(7):1929–1935. doi: 10.1128/jb.175.7.1929-1935.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binnie C., Lampe M., Losick R. Gene encoding the sigma 37 species of RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A. 1986 Aug;83(16):5943–5947. doi: 10.1073/pnas.83.16.5943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan S. A., Redfield A. R., Price C. W. Transcription factor sigma B of Bacillus subtilis controls a large stationary-phase regulon. J Bacteriol. 1993 Jul;175(13):3957–3963. doi: 10.1128/jb.175.13.3957-3963.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan S. A., Rutherford A., Thomas S. M., Price C. W. Activation of Bacillus subtilis transcription factor sigma B by a regulatory pathway responsive to stationary-phase signals. J Bacteriol. 1992 Jun;174(11):3695–3706. doi: 10.1128/jb.174.11.3695-3706.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boylan S. A., Thomas M. D., Price C. W. Genetic method to identify regulons controlled by nonessential elements: isolation of a gene dependent on alternate transcription factor sigma B of Bacillus subtilis. J Bacteriol. 1991 Dec;173(24):7856–7866. doi: 10.1128/jb.173.24.7856-7866.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burbulys D., Trach K. A., Hoch J. A. Initiation of sporulation in B. subtilis is controlled by a multicomponent phosphorelay. Cell. 1991 Feb 8;64(3):545–552. doi: 10.1016/0092-8674(91)90238-t. [DOI] [PubMed] [Google Scholar]
- Duncan L., Losick R. SpoIIAB is an anti-sigma factor that binds to and inhibits transcription by regulatory protein sigma F from Bacillus subtilis. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2325–2329. doi: 10.1073/pnas.90.6.2325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duncan M. L., Kalman S. S., Thomas S. M., Price C. W. Gene encoding the 37,000-dalton minor sigma factor of Bacillus subtilis RNA polymerase: isolation, nucleotide sequence, chromosomal locus, and cryptic function. J Bacteriol. 1987 Feb;169(2):771–778. doi: 10.1128/jb.169.2.771-778.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haldenwang W. G., Lang N., Losick R. A sporulation-induced sigma-like regulatory protein from B. subtilis. Cell. 1981 Feb;23(2):615–624. doi: 10.1016/0092-8674(81)90157-4. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Losick R. A modified RNA polymerase transcribes a cloned gene under sporulation control in Bacillus subtilis. Nature. 1979 Nov 15;282(5736):256–260. doi: 10.1038/282256a0. [DOI] [PubMed] [Google Scholar]
- Haldenwang W. G., Losick R. Novel RNA polymerase sigma factor from Bacillus subtilis. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7000–7004. doi: 10.1073/pnas.77.12.7000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igo M. M., Losick R. Regulation of a promoter that is utilized by minor forms of RNA polymerase holoenzyme in Bacillus subtilis. J Mol Biol. 1986 Oct 20;191(4):615–624. doi: 10.1016/0022-2836(86)90449-3. [DOI] [PubMed] [Google Scholar]
- Igo M., Lampe M., Ray C., Schafer W., Moran C. P., Jr, Losick R. Genetic studies of a secondary RNA polymerase sigma factor in Bacillus subtilis. J Bacteriol. 1987 Aug;169(8):3464–3469. doi: 10.1128/jb.169.8.3464-3469.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalman S., Duncan M. L., Thomas S. M., Price C. W. Similar organization of the sigB and spoIIA operons encoding alternate sigma factors of Bacillus subtilis RNA polymerase. J Bacteriol. 1990 Oct;172(10):5575–5585. doi: 10.1128/jb.172.10.5575-5585.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchman P. A., DeGrazia H., Kellner E. M., Moran C. P., Jr Forespore-specific disappearance of the sigma-factor antagonist spoIIAB: implications for its role in determination of cell fate in Bacillus subtilis. Mol Microbiol. 1993 May;8(4):663–671. doi: 10.1111/j.1365-2958.1993.tb01610.x. [DOI] [PubMed] [Google Scholar]
- Min K. T., Hilditch C. M., Diederich B., Errington J., Yudkin M. D. Sigma F, the first compartment-specific transcription factor of B. subtilis, is regulated by an anti-sigma factor that is also a protein kinase. Cell. 1993 Aug 27;74(4):735–742. doi: 10.1016/0092-8674(93)90520-z. [DOI] [PubMed] [Google Scholar]
- Rather P. N., Coppolecchia R., DeGrazia H., Moran C. P., Jr Negative regulator of sigma G-controlled gene expression in stationary-phase Bacillus subtilis. J Bacteriol. 1990 Feb;172(2):709–715. doi: 10.1128/jb.172.2.709-715.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt R., Margolis P., Duncan L., Coppolecchia R., Moran C. P., Jr, Losick R. Control of developmental transcription factor sigma F by sporulation regulatory proteins SpoIIAA and SpoIIAB in Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Dec;87(23):9221–9225. doi: 10.1073/pnas.87.23.9221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varón D., Boylan S. A., Okamoto K., Price C. W. Bacillus subtilis gtaB encodes UDP-glucose pyrophosphorylase and is controlled by stationary-phase transcription factor sigma B. J Bacteriol. 1993 Jul;175(13):3964–3971. doi: 10.1128/jb.175.13.3964-3971.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yansura D. G., Henner D. J. Use of the Escherichia coli lac repressor and operator to control gene expression in Bacillus subtilis. Proc Natl Acad Sci U S A. 1984 Jan;81(2):439–443. doi: 10.1073/pnas.81.2.439. [DOI] [PMC free article] [PubMed] [Google Scholar]