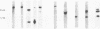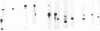Abstract
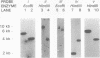
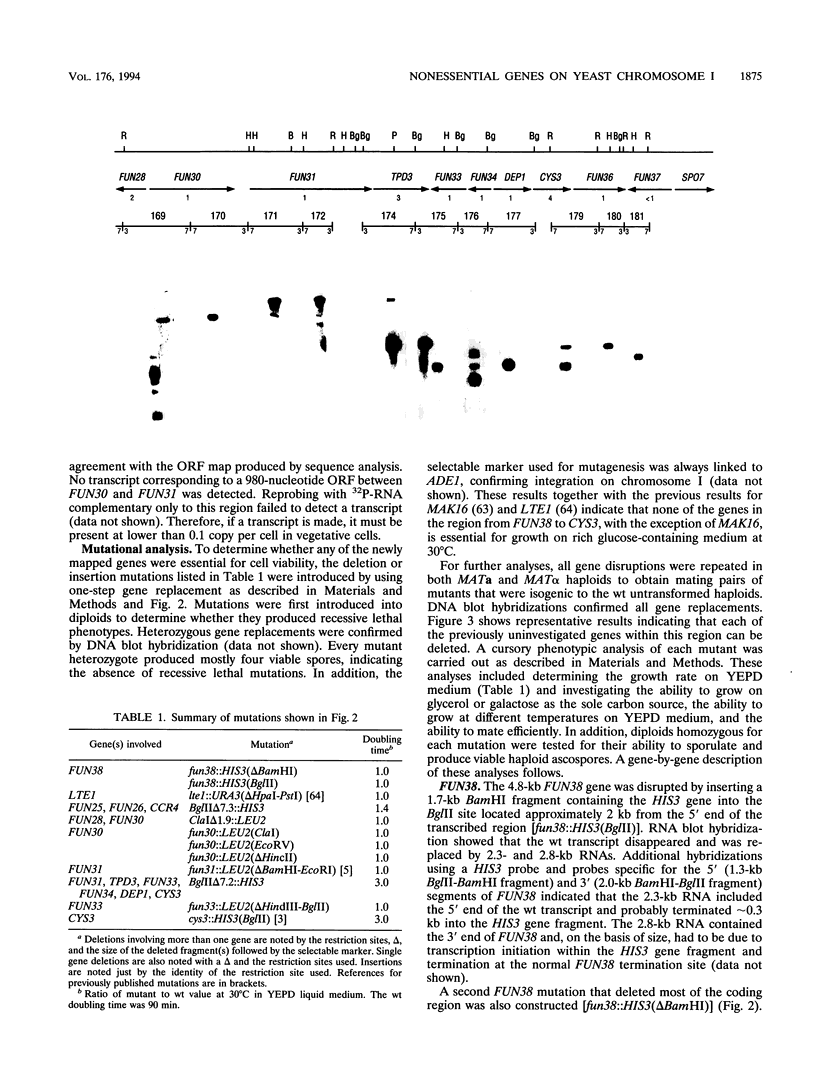
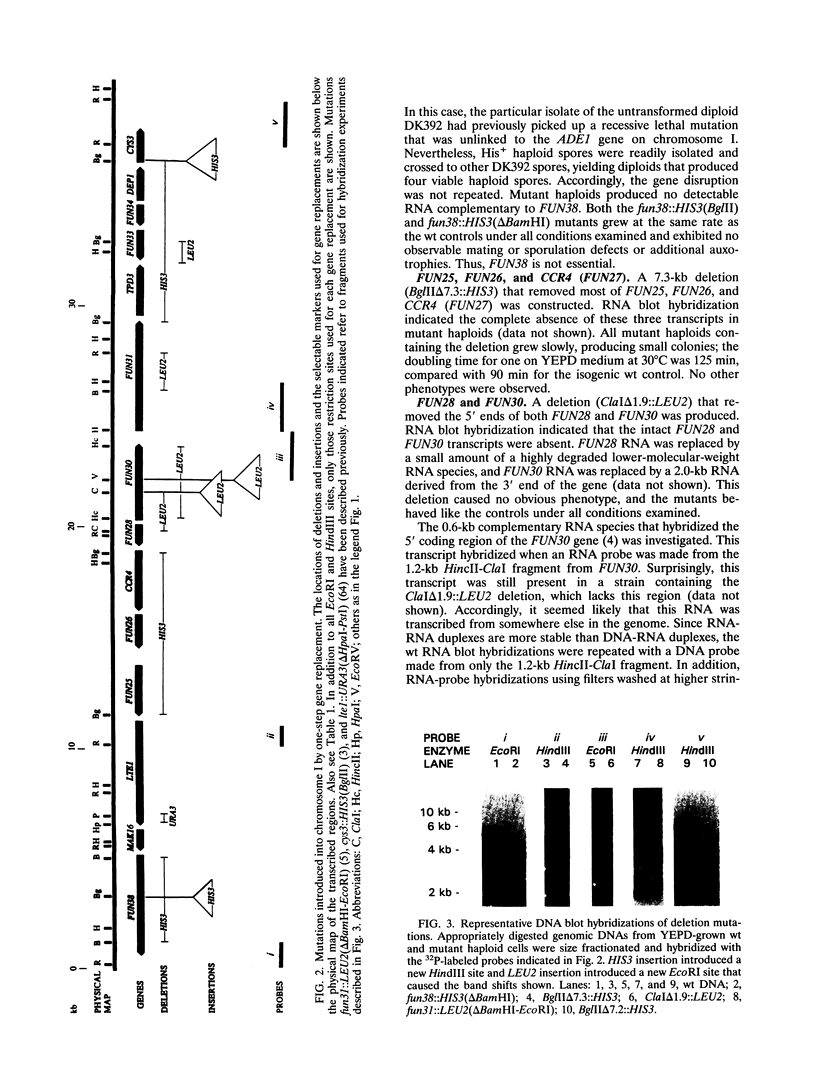
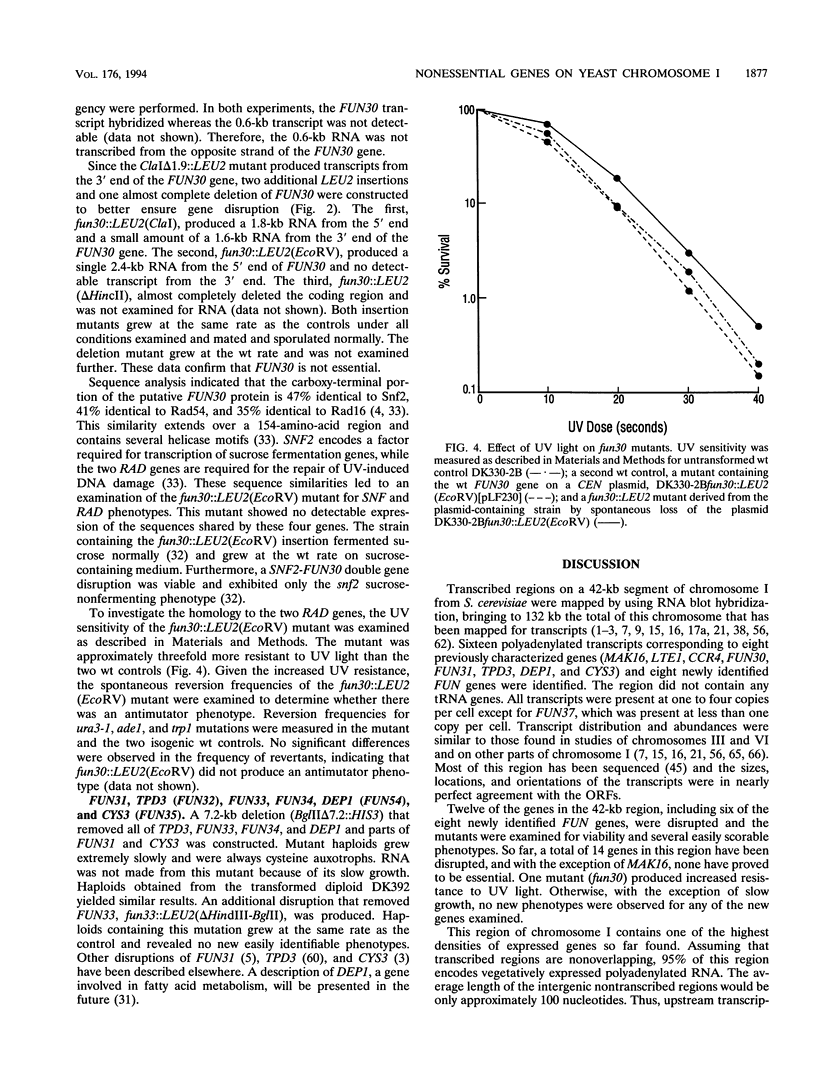
Transcribed regions on a 42-kb segment of chromosome I from Saccharomyces cerevisiae were mapped. Polyadenylated transcripts corresponding to eight previously characterized genes (MAK16, LTE1, CCR4, FUN30, FUN31, TPD3, DEP1, and CYS3) and eight new genes were identified. All transcripts were present at one to four copies per cell except for one which was significantly less abundant. This region has been sequenced, and the sizes, locations, and orientations of the transcripts were in nearly perfect agreement with the open reading frames. Disruptions in eight genes identified solely on the basis of a transcribed region, FUN38, FUN25, FUN26, FUN28, FUN30, FUN31, FUN33, and FUN34, indicated that all were nonessential for growth on rich medium at 30 degrees C. Disruption of FUN30, a gene closely related to RAD16 and RAD54, surprisingly resulted in increased resistance to UV irradiation. No additional phenotypes, other than slow growth, were observed for all other mutants. The distribution of essential genes on chromosome I is discussed.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barton A. B., Davies C. J., Hutchison C. A., 3rd, Kaback D. B. Cloning of chromosome I DNA from Saccharomyces cerevisiae: analysis of the FUN52 gene, whose product has homology to protein kinases. Gene. 1992 Aug 1;117(1):137–140. doi: 10.1016/0378-1119(92)90502-g. [DOI] [PubMed] [Google Scholar]
- Barton A. B., Kaback D. B., Clark M. W., Keng T., Ouellette B. F., Storms R. K., Zeng B., Zhong W., Fortin N., Delaney S. Physical localization of yeast CYS3, a gene whose product resembles the rat gamma-cystathionase and Escherichia coli cystathionine gamma-synthase enzymes. Yeast. 1993 Apr;9(4):363–369. doi: 10.1002/yea.320090406. [DOI] [PubMed] [Google Scholar]
- Clark M. W., Zhong W. W., Keng T., Storms R. K., Barton A., Kaback D. B., Bussey H. Identification of a Saccharomyces cerevisiae homolog of the SNF2 transcriptional regulator in the DNA sequence of an 8.6 kb region in the LTE1-CYS1 interval on the left arm of chromosome I. Yeast. 1992 Feb;8(2):133–145. doi: 10.1002/yea.320080208. [DOI] [PubMed] [Google Scholar]
- Clark M. W., Zhong W. W., Keng T., Storms R. K., Ouellette B. F., Barton A., Kaback D. B., Bussey H. The YAL017 gene on the left arm of chromosome I of Saccharomyces cerevisiae encodes a putative serine/threonine protein kinase. Yeast. 1993 May;9(5):543–549. doi: 10.1002/yea.320090511. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Helinski D. R. Effect of growth conditions on the formation of the relaxation complex of supercoiled ColE1 deoxyribonucleic acid and protein in Escherichia coli. J Bacteriol. 1972 Jun;110(3):1135–1146. doi: 10.1128/jb.110.3.1135-1146.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman K. G., Steensma H. Y., Kaback D. B., Pringle J. R. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation and characterization of the CDC24 gene and adjacent regions of the chromosome. Mol Cell Biol. 1986 Dec;6(12):4516–4525. doi: 10.1128/mcb.6.12.4516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross F. R. DAF1, a mutant gene affecting size control, pheromone arrest, and cell cycle kinetics of Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4675–4684. doi: 10.1128/mcb.8.11.4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crowley J. C., Kaback D. B. Cloning of chromosome I DNA from Saccharomyces cerevisiae: mutational analysis of the FUN2 transcribed region. Gene. 1989 Nov 30;83(2):381–385. doi: 10.1016/0378-1119(89)90126-1. [DOI] [PubMed] [Google Scholar]
- Crowley J. C., Kaback D. B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation of the ADE1 gene. J Bacteriol. 1984 Jul;159(1):413–417. doi: 10.1128/jb.159.1.413-417.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummins C. M., Culbertson M. R., Knapp G. Frameshift suppressor mutations outside the anticodon in yeast proline tRNAs containing an intervening sequence. Mol Cell Biol. 1985 Jul;5(7):1760–1771. doi: 10.1128/mcb.5.7.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Virgilio C., Bürckert N., Barth G., Neuhaus J. M., Boller T., Wiemken A. Cloning and disruption of a gene required for growth on acetate but not on ethanol: the acetyl-coenzyme A synthetase gene of Saccharomyces cerevisiae. Yeast. 1992 Dec;8(12):1043–1051. doi: 10.1002/yea.320081207. [DOI] [PubMed] [Google Scholar]
- Diehl B. E., Pringle J. R. Molecular analysis of Saccharomyces cerevisiae chromosome I: identification of additional transcribed regions and demonstration that some encode essential functions. Genetics. 1991 Feb;127(2):287–298. doi: 10.1093/genetics/127.2.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerst J. E., Rodgers L., Riggs M., Wigler M. SNC1, a yeast homolog of the synaptic vesicle-associated membrane protein/synaptobrevin gene family: genetic interactions with the RAS and CAP genes. Proc Natl Acad Sci U S A. 1992 May 15;89(10):4338–4342. doi: 10.1073/pnas.89.10.4338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebl M. G., Petes T. D. Most of the yeast genomic sequences are not essential for cell growth and division. Cell. 1986 Sep 26;46(7):983–992. doi: 10.1016/0092-8674(86)90697-5. [DOI] [PubMed] [Google Scholar]
- Gottlin-Ninfa E., Kaback D. B. Isolation and functional analysis of sporulation-induced transcribed sequences from Saccharomyces cerevisiae. Mol Cell Biol. 1986 Jun;6(6):2185–2197. doi: 10.1128/mcb.6.6.2185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn S., Pinkham J., Wei R., Miller R., Guarente L. The HAP3 regulatory locus of Saccharomyces cerevisiae encodes divergent overlapping transcripts. Mol Cell Biol. 1988 Feb;8(2):655–663. doi: 10.1128/mcb.8.2.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris S. D., Cheng J., Pugh T. A., Pringle J. R. Molecular analysis of Saccharomyces cerevisiae chromosome I. On the number of genes and the identification of essential genes using temperature-sensitive-lethal mutations. J Mol Biol. 1992 May 5;225(1):53–65. doi: 10.1016/0022-2836(92)91025-k. [DOI] [PubMed] [Google Scholar]
- Harris S. D., Pringle J. R. Genetic analysis of Saccharomyces cerevisiae chromosome I: on the role of mutagen specificity in delimiting the set of genes identifiable using temperature-sensitive-lethal mutations. Genetics. 1991 Feb;127(2):279–285. doi: 10.1093/genetics/127.2.279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W. D., Rao M. R., Erdile L. F., Kelly T. J., Kolodner R. D. An essential Saccharomyces cerevisiae single-stranded DNA binding protein is homologous to the large subunit of human RP-A. EMBO J. 1990 Jul;9(7):2321–2329. doi: 10.1002/j.1460-2075.1990.tb07404.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffman C. S., Winston F. A ten-minute DNA preparation from yeast efficiently releases autonomous plasmids for transformation of Escherichia coli. Gene. 1987;57(2-3):267–272. doi: 10.1016/0378-1119(87)90131-4. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback D. B., Oeller P. W., Yde Steensma H., Hirschman J., Ruezinsky D., Coleman K. G., Pringle J. R. Temperature-sensitive lethal mutations on yeast chromosome I appear to define only a small number of genes. Genetics. 1984 Sep;108(1):67–90. doi: 10.1093/genetics/108.1.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaback D. B., Steensma H. Y., de Jonge P. Enhanced meiotic recombination on the smallest chromosome of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1989 May;86(10):3694–3698. doi: 10.1073/pnas.86.10.3694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Küenzi M. T., Tingle M. A., Halvorson H. O. Sporulation of Saccharomyces cerevisiae in the absence of a functional mitochondrial genome. J Bacteriol. 1974 Jan;117(1):80–88. doi: 10.1128/jb.117.1.80-88.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurent B. C., Yang X., Carlson M. An essential Saccharomyces cerevisiae gene homologous to SNF2 encodes a helicase-related protein in a new family. Mol Cell Biol. 1992 Apr;12(4):1893–1902. doi: 10.1128/mcb.12.4.1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre O., Carles C., Conesa C., Swanson R. N., Bouet F., Riva M., Sentenac A. TFC3: gene encoding the B-block binding subunit of the yeast transcription factor IIIC. Proc Natl Acad Sci U S A. 1992 Nov 1;89(21):10512–10516. doi: 10.1073/pnas.89.21.10512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malavasic M. J., Elder R. T. Complementary transcripts from two genes necessary for normal meiosis in the yeast Saccharomyces cerevisiae. Mol Cell Biol. 1990 Jun;10(6):2809–2819. doi: 10.1128/mcb.10.6.2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvar T., Biron R. W., Kaback D. B., Denis C. L. The CCR4 protein from Saccharomyces cerevisiae contains a leucine-rich repeat region which is required for its control of ADH2 gene expression. Genetics. 1992 Dec;132(4):951–962. doi: 10.1093/genetics/132.4.951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller P. P., Hinnebusch A. G. Multiple upstream AUG codons mediate translational control of GCN4. Cell. 1986 Apr 25;45(2):201–207. doi: 10.1016/0092-8674(86)90384-3. [DOI] [PubMed] [Google Scholar]
- Nash R., Tokiwa G., Anand S., Erickson K., Futcher A. B. The WHI1+ gene of Saccharomyces cerevisiae tethers cell division to cell size and is a cyclin homolog. EMBO J. 1988 Dec 20;7(13):4335–4346. doi: 10.1002/j.1460-2075.1988.tb03332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman R. B., Kaback D. B., Dubin R. A., Perkins E. L., Rosenberg N. G., Sutherland K. A., Forrest D. B., Michels C. A. MAL6 of Saccharomyces: a complex genetic locus containing three genes required for maltose fermentation. Proc Natl Acad Sci U S A. 1984 May;81(9):2811–2815. doi: 10.1073/pnas.81.9.2811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng R., Abelson J. Isolation and sequence of the gene for actin in Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1980 Jul;77(7):3912–3916. doi: 10.1073/pnas.77.7.3912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Connor J. P., Peebles C. L. PTA1, an essential gene of Saccharomyces cerevisiae affecting pre-tRNA processing. Mol Cell Biol. 1992 Sep;12(9):3843–3856. doi: 10.1128/mcb.12.9.3843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohmen J. D., Burke K. A., McEwen J. E. Divergent overlapping transcripts at the PET122 locus in Saccharomyces cerevisiae. Mol Cell Biol. 1990 Jun;10(6):3027–3035. doi: 10.1128/mcb.10.6.3027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Ono B., Tanaka K., Naito K., Heike C., Shinoda S., Yamamoto S., Ohmori S., Oshima T., Toh-e A. Cloning and characterization of the CYS3 (CYI1) gene of Saccharomyces cerevisiae. J Bacteriol. 1992 May;174(10):3339–3347. doi: 10.1128/jb.174.10.3339-3347.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouellette B. F., Clark M. W., Keng T., Storms R. K., Zhong W., Zeng B., Fortin N., Delaney S., Barton A., Kaback D. B. Sequencing of chromosome I from Saccharomyces cerevisiae: analysis of a 32 kb region between the LTE1 and SPO7 genes. Genome. 1993 Feb;36(1):32–42. doi: 10.1139/g93-005. [DOI] [PubMed] [Google Scholar]
- Riles L., Dutchik J. E., Baktha A., McCauley B. K., Thayer E. C., Leckie M. P., Braden V. V., Depke J. E., Olson M. V. Physical maps of the six smallest chromosomes of Saccharomyces cerevisiae at a resolution of 2.6 kilobase pairs. Genetics. 1993 May;134(1):81–150. doi: 10.1093/genetics/134.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riles L., Olson M. V. Nonsense mutations in essential genes of Saccharomyces cerevisiae. Genetics. 1988 Apr;118(4):601–607. doi: 10.1093/genetics/118.4.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothstein R. J. One-step gene disruption in yeast. Methods Enzymol. 1983;101:202–211. doi: 10.1016/0076-6879(83)01015-0. [DOI] [PubMed] [Google Scholar]
- Schweitzer B., Philippsen P. CDC15, an essential cell cycle gene in Saccharomyces cerevisiae, encodes a protein kinase domain. Yeast. 1991 Apr;7(3):265–273. doi: 10.1002/yea.320070308. [DOI] [PubMed] [Google Scholar]
- Sikorski R. S., Hieter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989 May;122(1):19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slater M. R., Craig E. A. The SSA1 and SSA2 genes of the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1989 Jan 25;17(2):805–806. doi: 10.1093/nar/17.2.805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Southern E. M. Detection of specific sequences among DNA fragments separated by gel electrophoresis. J Mol Biol. 1975 Nov 5;98(3):503–517. doi: 10.1016/s0022-2836(75)80083-0. [DOI] [PubMed] [Google Scholar]
- Steensma H. Y., Crowley J. C., Kaback D. B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation and analysis of the CEN1-ADE1-CDC15 region. Mol Cell Biol. 1987 Jan;7(1):410–419. doi: 10.1128/mcb.7.1.410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka S., Yoshikawa A., Isono K. An ordered clone bank for chromosome I of Saccharomyces cerevisiae. J Bacteriol. 1992 Sep;174(18):5985–5987. doi: 10.1128/jb.174.18.5985-5987.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teunissen A. W., Holub E., van der Hucht J., van den Berg J. A., Steensma H. Y. Sequence of the open reading frame of the FLO1 gene from Saccharomyces cerevisiae. Yeast. 1993 Apr;9(4):423–427. doi: 10.1002/yea.320090413. [DOI] [PubMed] [Google Scholar]
- Thomas P. S. Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc Natl Acad Sci U S A. 1980 Sep;77(9):5201–5205. doi: 10.1073/pnas.77.9.5201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Venter U., Hörz W. The acid phosphatase genes PHO10 and PHO11 in S. cerevisiae are located at the telomeres of chromosomes VIII and I. Nucleic Acids Res. 1989 Feb 25;17(4):1353–1369. doi: 10.1093/nar/17.4.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whyte W., Keopp L. H., Lamb J., Crowley J. C., Kaback D. B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation, characterization and regulation of the SPO7 sporulation gene. Gene. 1990 Oct 30;95(1):65–72. doi: 10.1016/0378-1119(90)90414-m. [DOI] [PubMed] [Google Scholar]
- Wickner R. B. Host function of MAK16: G1 arrest by a mak16 mutant of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1988 Aug;85(16):6007–6011. doi: 10.1073/pnas.85.16.6007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickner R. B., Koh T. J., Crowley J. C., O'Neil J., Kaback D. B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: isolation of the MAK16 gene and analysis of an adjacent gene essential for growth at low temperatures. Yeast. 1987 Mar;3(1):51–57. doi: 10.1002/yea.320030108. [DOI] [PubMed] [Google Scholar]
- Yoshikawa A., Isono K. Chromosome III of Saccharomyces cerevisiae: an ordered clone bank, a detailed restriction map and analysis of transcripts suggest the presence of 160 genes. Yeast. 1990 Sep-Oct;6(5):383–401. doi: 10.1002/yea.320060504. [DOI] [PubMed] [Google Scholar]
- Yoshikawa A., Isono K. Construction of an ordered clone bank and systematic analysis of the whole transcripts of chromosome VI of Saccharomyces cerevisiae. Nucleic Acids Res. 1991 Mar 25;19(6):1189–1195. doi: 10.1093/nar/19.6.1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Steensma H. Y., de Jonge P., Kaptein A., Kaback D. B. Molecular cloning of chromosome I DNA from Saccharomyces cerevisiae: localization of a repeated sequence containing an acid phosphatase gene near a telomere of chromosome I and chromosome VIII. Curr Genet. 1989 Sep;16(3):131–137. doi: 10.1007/BF00391468. [DOI] [PubMed] [Google Scholar]
- de Virgilio C., Bürckert N., Neuhaus J. M., Boller T., Wiemken A. CNE1, a Saccharomyces cerevisiae homologue of the genes encoding mammalian calnexin and calreticulin. Yeast. 1993 Feb;9(2):185–188. doi: 10.1002/yea.320090209. [DOI] [PubMed] [Google Scholar]
- van Zyl W., Huang W., Sneddon A. A., Stark M., Camier S., Werner M., Marck C., Sentenac A., Broach J. R. Inactivation of the protein phosphatase 2A regulatory subunit A results in morphological and transcriptional defects in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Nov;12(11):4946–4959. doi: 10.1128/mcb.12.11.4946. [DOI] [PMC free article] [PubMed] [Google Scholar]