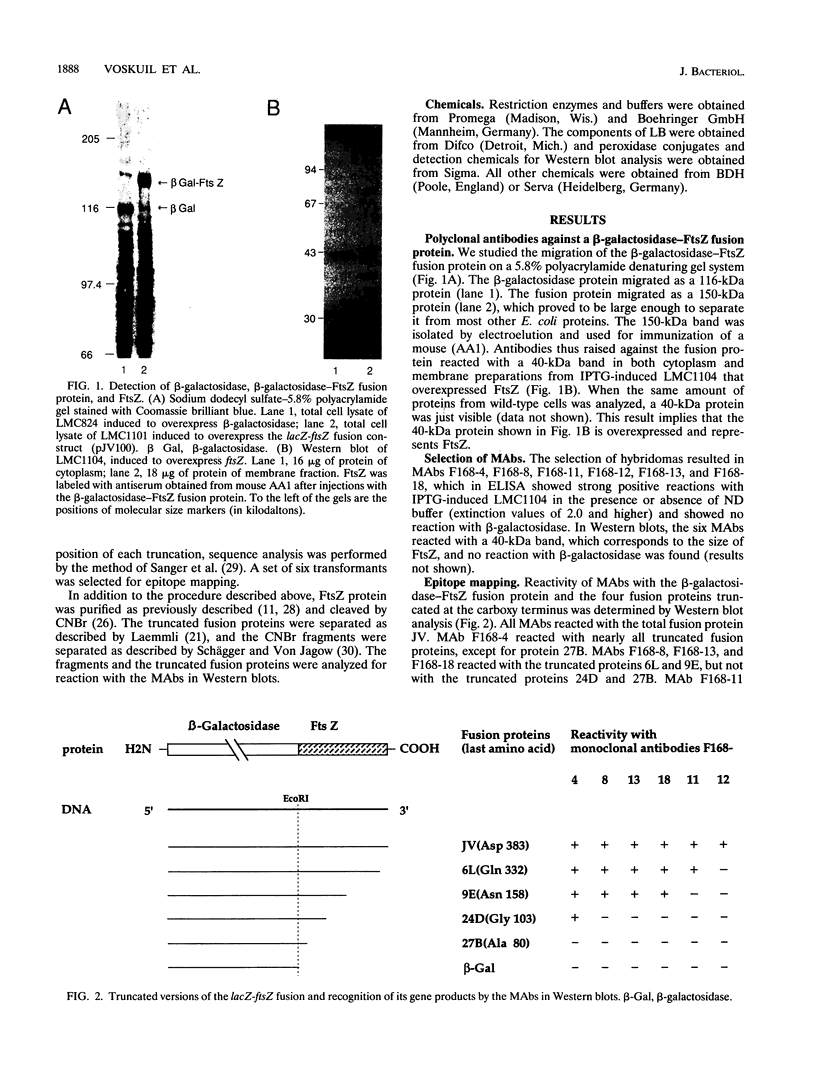
Abstract
A fusion between lacZ and ftsZ of Escherichia coli was constructed to obtain a beta-galactosidase-FtsZ fusion protein. This fusion protein was used to raise antibodies against cell division protein FtsZ. Six monoclonal antibodies were obtained, and they reacted with FtsZ from cytoplasm and membrane fractions. The epitopes in FtsZ were localized by studying the reactions of the monoclonal antibodies with fusion proteins truncated at the carboxy terminus and with fragments that were obtained by CNBr cleavage of purified FtsZ. Five different epitopes were defined. Epitopes I and III reacted with the same monoclonal antibody, without showing apparent amino acid homology. Epitope II was defined by monoclonal antibodies that cross-reacted with an unknown cytoplasmic 50-kDa protein not related to FtsZ. Epitopes IV and V were recognized by different monoclonal antibodies. All monoclonal antibodies reacted strongly under native conditions, so it is likely that the five epitopes are situated on the surface of native FtsZ. By using these data and computer analysis, a provisional model of FtsZ is proposed. The FtsZ protein is considered to be globular, with a hydrophobic pocket containing GTP-binding elements. Epitopes I and II are situated on each side of the hydrophobic pocket. Because the carboxy terminus contains epitope V, the carboxy terminus of FtsZ is likely oriented toward the protein's surface.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aldea M., Garrido T., Pla J., Vicente M. Division genes in Escherichia coli are expressed coordinately to cell septum requirements by gearbox promoters. EMBO J. 1990 Nov;9(11):3787–3794. doi: 10.1002/j.1460-2075.1990.tb07592.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Begg K. J., Donachie W. D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985 Aug;163(2):615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. F., Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991 Nov 14;354(6349):161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA). J Bacteriol. 1990 Oct;172(10):5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourne H. R., Sanders D. A., McCormick F. The GTPase superfamily: conserved structure and molecular mechanism. Nature. 1991 Jan 10;349(6305):117–127. doi: 10.1038/349117a0. [DOI] [PubMed] [Google Scholar]
- Casadaban M. J. Transposition and fusion of the lac genes to selected promoters in Escherichia coli using bacteriophage lambda and Mu. J Mol Biol. 1976 Jul 5;104(3):541–555. doi: 10.1016/0022-2836(76)90119-4. [DOI] [PubMed] [Google Scholar]
- Chung C. T., Miller R. H. A rapid and convenient method for the preparation and storage of competent bacterial cells. Nucleic Acids Res. 1988 Apr 25;16(8):3580–3580. doi: 10.1093/nar/16.8.3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K., Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992 Oct;174(19):6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K., Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991 Jun;173(11):3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar S. J., Begg K. J., Donachie W. D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992 Oct;174(19):6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar S. J., Kagan-Zur V., Begg K. J., Donachie W. D. Transcriptional regulation of cell division genes in Escherichia coli. Mol Microbiol. 1989 Oct;3(10):1371–1377. doi: 10.1111/j.1365-2958.1989.tb00118.x. [DOI] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Garrido T., Sánchez M., Palacios P., Aldea M., Vicente M. Transcription of ftsZ oscillates during the cell cycle of Escherichia coli. EMBO J. 1993 Oct;12(10):3957–3965. doi: 10.1002/j.1460-2075.1993.tb06073.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huisman O., D'Ari R., Gottesman S. Cell-division control in Escherichia coli: specific induction of the SOS function SfiA protein is sufficient to block septation. Proc Natl Acad Sci U S A. 1984 Jul;81(14):4490–4494. doi: 10.1073/pnas.81.14.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk A. H., Evers R., Groothuis D. G., Gilis H., Kuijper S. Production and characterization of monoclonal antibodies against specific serotypes of Mycobacterium avium and the Mycobacterium avium-Mycobacterium intracellulare-Mycobacterium scrofulaceum complex. Infect Immun. 1989 Aug;57(8):2514–2521. doi: 10.1128/iai.57.8.2514-2521.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolk A. H., Ho M. L., Klatser P. R., Eggelte T. A., Kuijper S., de Jonge S., van Leeuwen J. Production and characterization of monoclonal antibodies to Mycobacterium tuberculosis, M. bovis (BCG) and M. leprae. Clin Exp Immunol. 1984 Dec;58(3):511–521. [PMC free article] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Köhler G., Hengartner H., Shulman M. J. Immunoglobulin production by lymphocyte hybridomas. Eur J Immunol. 1978 Feb;8(2):82–88. doi: 10.1002/eji.1830080203. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lutkenhaus J. FtsZ ring in bacterial cytokinesis. Mol Microbiol. 1993 Aug;9(3):403–409. doi: 10.1111/j.1365-2958.1993.tb01701.x. [DOI] [PubMed] [Google Scholar]
- Masters M., Paterson T., Popplewell A. G., Owen-Hughes T., Pringle J. H., Begg K. J. The effect of DnaA protein levels and the rate of initiation at oriC on transcription originating in the ftsQ and ftsA genes: in vivo experiments. Mol Gen Genet. 1989 Apr;216(2-3):475–483. doi: 10.1007/BF00334393. [DOI] [PubMed] [Google Scholar]
- Mukherjee A., Dai K., Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanninga N. Cell division and peptidoglycan assembly in Escherichia coli. Mol Microbiol. 1991 Apr;5(4):791–795. doi: 10.1111/j.1365-2958.1991.tb00751.x. [DOI] [PubMed] [Google Scholar]
- Nikodem V., Fresco J. R. Protein fingerprinting by SDS-gel electrophoresis after partial fragmentation with CNBr. Anal Biochem. 1979 Sep 1;97(2):382–386. doi: 10.1016/0003-2697(79)90089-7. [DOI] [PubMed] [Google Scholar]
- Pla J., Sánchez M., Palacios P., Vicente M., Aldea M. Preferential cytoplasmic location of FtsZ, a protein essential for Escherichia coli septation. Mol Microbiol. 1991 Jul;5(7):1681–1686. doi: 10.1111/j.1365-2958.1991.tb01915.x. [DOI] [PubMed] [Google Scholar]
- RayChaudhuri D., Park J. T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992 Sep 17;359(6392):251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schägger H., von Jagow G. Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal Biochem. 1987 Nov 1;166(2):368–379. doi: 10.1016/0003-2697(87)90587-2. [DOI] [PubMed] [Google Scholar]
- Smith R. W., Masters M., Donachie W. D. Cell division and transcription of ftsZ. J Bacteriol. 1993 May;175(9):2788–2791. doi: 10.1128/jb.175.9.2788-2791.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stanley K. K., Luzio J. P. Construction of a new family of high efficiency bacterial expression vectors: identification of cDNA clones coding for human liver proteins. EMBO J. 1984 Jun;3(6):1429–1434. doi: 10.1002/j.1460-2075.1984.tb01988.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taschner P. E., Huls P. G., Pas E., Woldringh C. L. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J Bacteriol. 1988 Apr;170(4):1533–1540. doi: 10.1128/jb.170.4.1533-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tétart F., Albigot R., Conter A., Mulder E., Bouché J. P. Involvement of FtsZ in coupling of nucleoid separation with septation. Mol Microbiol. 1992 Mar;6(5):621–627. doi: 10.1111/j.1365-2958.1992.tb01509.x. [DOI] [PubMed] [Google Scholar]
- Tétart F., Bouché J. P. Regulation of the expression of the cell-cycle gene ftsZ by DicF antisense RNA. Division does not require a fixed number of FtsZ molecules. Mol Microbiol. 1992 Mar;6(5):615–620. doi: 10.1111/j.1365-2958.1992.tb01508.x. [DOI] [PubMed] [Google Scholar]
- Verbon A., Kuijper S., Jansen H. M., Speelman P., Kolk A. H. Antigens in culture supernatant of Mycobacterium tuberculosis: epitopes defined by monoclonal and human antibodies. J Gen Microbiol. 1990 May;136(5):955–964. doi: 10.1099/00221287-136-5-955. [DOI] [PubMed] [Google Scholar]
- Vicente M., Kushner S. R., Garrido T., Aldea M. The role of the 'gearbox' in the transcription of essential genes. Mol Microbiol. 1991 Sep;5(9):2085–2091. doi: 10.1111/j.1365-2958.1991.tb02137.x. [DOI] [PubMed] [Google Scholar]
- Wang X. D., de Boer P. A., Rothfield L. I. A factor that positively regulates cell division by activating transcription of the major cluster of essential cell division genes of Escherichia coli. EMBO J. 1991 Nov;10(11):3363–3372. doi: 10.1002/j.1460-2075.1991.tb04900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. E., Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985 Oct;42(3):941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- Wientjes F. B., Nanninga N. Rate and topography of peptidoglycan synthesis during cell division in Escherichia coli: concept of a leading edge. J Bacteriol. 1989 Jun;171(6):3412–3419. doi: 10.1128/jb.171.6.3412-3419.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh C. L., Mulder E., Huls P. G., Vischer N. Toporegulation of bacterial division according to the nucleoid occlusion model. Res Microbiol. 1991 Feb-Apr;142(2-3):309–320. doi: 10.1016/0923-2508(91)90046-d. [DOI] [PubMed] [Google Scholar]
- Yi Q. M., Lutkenhaus J. The nucleotide sequence of the essential cell-division gene ftsZ of Escherichia coli. Gene. 1985;36(3):241–247. doi: 10.1016/0378-1119(85)90179-9. [DOI] [PubMed] [Google Scholar]
- de Boer P., Crossley R., Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992 Sep 17;359(6392):254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]







