SUMMARY
Microtubule plus end binding proteins (+TIPS) localize to the dynamic plus ends of microtubules where they stimulate microtubule growth and recruit signaling molecules. Three main +TIP classes have been identified (XMAP215, EB1 and CLIP-170), but whether they act upon microtubule plus ends through a similar mechanism has not been resolved. Here, we report crystal structures of the tubulin binding domains of XMAP215 (yeast Stu2p and Drosophila Msps), EB1 (yeast Bim1p and human EB1), and CLIP-170 (human), which reveal diverse tubulin binding interfaces. Functional studies, however, reveal a common property that native or artificial dimerization of tubulin binding domains (including chemically-induced heterodimers of EB1 and CLIP-170) induces tubulin nucleation/assembly in vitro and, in most cases, plus end tracking in living cells. We propose that +TIPs, although diverse in structure, share a common property of multimerizing tubulin, thus acting as polymerization chaperones that aid in subunit addition to the microtubule plus end.
INTRODUCTION
Microtubules are highly dynamic polymers that are utilized for intracellular transport, the construction of the mitotic spindle, and spatial organization (e.g. polarity) of eukaryotic cells. Microtubules, cylindrical polymers of 13 protofilaments, are inherently asymmetric owing to the head-to-tail polymerization of the αβ tubulin heterodimer, which propagates at a supermolecular level to create distinct ‘minus’ and ‘plus’ ends of the microtubule. In most cells, microtubule polymerization is initiated at specific locations (e.g. the centrosome), generating microtubule arrays of fixed polarity with the plus ends extending away from the sites of nucleation. Much of the cellular microtubule dynamics (transitions between growth and shrinkage of polymer) occurs at the microtubule plus end. Growing and shrinking plus ends enable microtubules to explore the cytoplasm, searching to make contacts with chromosomes during mitosis or engaging the cell cortex to direct motility in migrating cells or growth cones (Lansbergen and Akhmanova, 2006).
Microtubules dynamics have been reconstituted with purified tubulin and GTP (Desai and Mitchison, 1997; Horio and Hotani, 1986; Mitchison and Kirschner, 1984). However, microtubules in cells exhibit distinct dynamic properties compared with pure tubulin, in some cases demonstrating more frequent interconversions between growth and shrinkage and in other cases, extreme stability with little change in microtubule length (Srayko et al., 2005; Tournebize et al., 2000). Such differences between the behavior of microtubules composed of pure tubulin and microtubules in cells is due to the actions of numerous microtubule or tubulin associated factors, some of which destabilize (e.g. Kinesin-13, stathmin/OP18, and katanin) while others stabilize (EB1 and neuronal MAPs) the microtubule (Kinoshita et al., 2001; McNally and Vale, 1993; Moores and Milligan, 2006; Rogers et al., 2002; Samsonov et al., 2004; Tournebize et al., 2000).
A particularly interesting class of microtubule binding proteins is the +TIPs, which associate selectively with the growing plus ends of microtubules. The first discovered +TIP was CLIP-170, which was found to localize to microtubule tips by immunofluorescence (Perez et al., 1999; Pierre et al., 1992). Subsequent live cell imaging revealed that CLIP-170-GFP tracked along growing microtubule plus ends, appearing as short fluorescent “comets” traveling centripetally through the cytoplasm and disappearing abruptly when the microtubule converted to a depolymerizing state (Perez et al., 1999; Pierre et al., 1992). Subsequently, the EB1 protein family and XMAP215 family were identified as +TIP proteins (Mimori-Kiyosue et al., 2000; van Breugel et al., 2003). CLIP-170 (and the related p150Glued protein of dynactin), EB1 (and the related CLAMP protein), and XMAP215 (and the related CLASP proteins (see Discussion)) each have distinct, characteristic domains and are found in virtually all eukaryotes. The roles of these proteins are numerous and complex. Loss or inhibition of these proteins compromises microtubule growth in many cells types (Brittle and Ohkura, 2005; Cullen et al., 1999; Rogers et al., 2002), and in some cases, enhanced nucleation and polymerization of microtubules has been demonstrated for purified +TIPs in reconstituted assays (Gard and Kirschner, 1987; Kerssemakers et al., 2006; Kinoshita et al., 2001; Vasquez et al., 1994). In addition, the +TIPs create a network of complex protein-protein interactions, including interactions between +TIPs themselves as well as with a second tier of signaling proteins and actin-binding proteins (Lansbergen and Akhmanova, 2006). In addition, +TIPS interact with a number of adaptor proteins, affording specific subcellular localization. For example, the XMAP215 family is specifically recruited to the centrosome by the TACC family of proteins indicative of +TIP functions beyond microtubule plus end regulation (Lee et al., 2001).
While the cell biological roles of +TIPs have been the subject of extensive studies in the last few years, the detailed mechanisms of how they localize to the plus end and affect microtubule dynamics remains to be resolved. Association of some +TIP domains with purified tubulin has suggested a model in which +TIPs bind tubulin monomers or oligomers in solution and then co-assemble onto the growing microtubule plus end (Diamantopoulos et al., 1999; Folker et al., 2005). However, in other cases, motor-driven transport of +TIPs to microtubule ends has been observed and suggested as another mechanism for plus end accumulation (Busch et al., 2004; Carvalho et al., 2004). The dissociation of +TIPs has been suggested to occur either by decreased affinity after incorporation into the mature microtubule lattice or by release due to phosphorylation (Tirnauer et al., 2004; Vaughan et al., 2002; Wittmann and Waterman-Storer, 2005).
In this study, we sought to compare the atomic structures and biochemical mechanisms of representatives of the three main +TIPs families: XMAP215 (also known by the S. pombe and human counterparts, Dis1 and Ch-TOG), CLIP-170 and EB1. We find each domain to be structurally unique with a conserved face utilized for tubulin association. However, single domains fail to promote microtubule growth in vitro and fail to engage in robust microtubule plus end tracking activity in vivo. Instead, dimerization of domains, either from within a single +TIP family or as heterodimerized domains across +TIP families, promote microtubule growth below tubulin’s critical concentration in vitro and restores plus end tracking activity in vivo. The studies reveal a common mechanism in which diverse tubulin binding scaffolds in +TIPs serve to oligomerize tubulin prior to loading onto the microtubule plus end to facilitate microtubule assembly.
RESULTS
Tubulin Binding and Polymerization Activity of +TIP Domains
Tubulin Binding by Gel Filtration
Prior to initiating structural studies, we first sought to express and define regions of +TIP proteins that interact with tubulin. For the XMAP215 family, sequence homology reveals a single common domain (termed the TOG domain) that is conserved across species (Andrade et al., 2001). This domain is repeated twice in the S. cerevisiae homolog Stu2p, while higher eukaryotes have five arrayed TOG domains (Figure 1A). Sequence analysis reveals that TOG domains can be further subclassified into two or possibly three types (termed A, B, and C TOG domains)(Figure S2). The TOG domain types alternate in the polypeptide: for higher eukaryotes TOG domains 1 and 3 are type A, TOG domains 2 and 4 are type B, and TOG domain 5 is type C (mostly closely related to type A). The predicted pI of TOG domain A is negative at physiological pH, while type B displays a net positive charge. C-terminal to the TOG domains, yeast Stu2p has a predicted coiled coil followed by a basic region that has been shown to bind to microtubules (Nakaseko et al., 2001), while higher eukaryotic homologues have an unique conserved C-terminal domain also implicated in microtubule association (Popov et al., 2001). In contrast to Stu2p which dimerizes, higher eukaryotic family members are monomeric (Al-Bassam et al., 2006; Graf et al., 2000; van Breugel et al., 2003).
Figure 1. Delineation of tubulin binding domains.
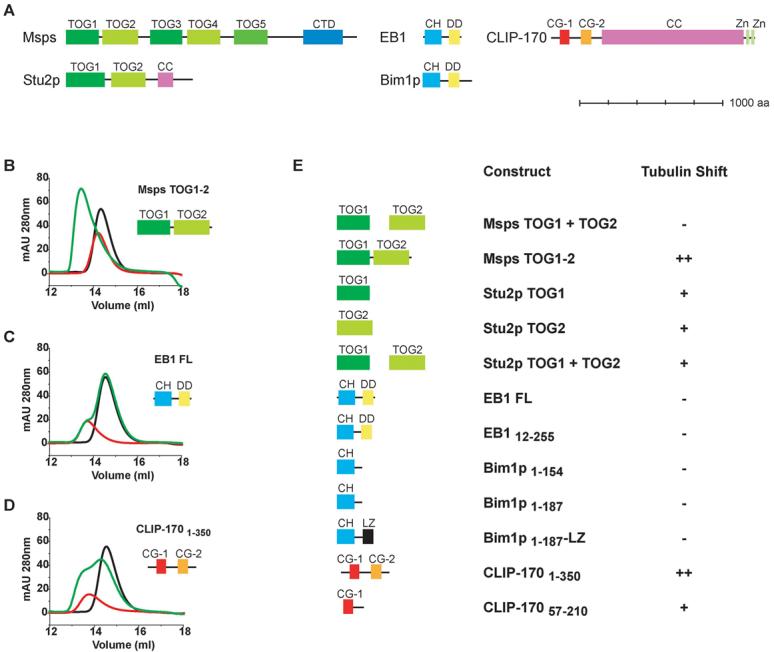
A) Domain structure diagrams of XMAP215 family members Msps and Stu2p, EB1 family members EB1 and Bim1p and CLIP-170. Domains indicated are TOG domains, CTD (C-Terminal Domain), CC (Coiled Coil), CH (Calponin Homology domain), DD (Dimerization Domain), CG (Cap-Gly) domain, Zn (Zinc-Finger Domain). Scale bar indicates 1000 amino acids. Gel filtrations studies testing the interaction of +TIP domains with tubulin (B-E). Traces are colored as: tubulin alone, black; +TIP domain(s) alone, red; tubulin plus +TIP, green. Plots indicate absorption at 280 nm on the y-axis and elution volume in ml along the x-axis. B) Msps TOG1-2. C) EB1 full length. D) CLIP-1701-350. E) Summary of gel filtration data presented in B-D and Supplementary fig. 1. where ++, + and - indicate strong, moderate and no tubulin shift respectively.
We expressed the TOG1 and 2 domains from the Drosophila XMAP215 homolog (termed Mini spindles, Msps) and yeast Stu2p and tested whether they formed a complex with α/β tubulin heterodimers by co-elution in gel filtration chromatography. Neither Msps TOG1 or TOG2 alone or added in trans formed a complex with tubulin, although a tandem TOG1-2 construct interacted with tubulin in this assay (Figures 1B and E; Figures S1A and B) corroborating preexisting evidence of an N-terminal tubulin binding domain in the XMAP215 family (Spittle et al., 2000). Analysis of the shifted TOG1-2:tubulin peak via gel filtration and dynamic light scattering indicated a mass of 121 kDa, which is less than the expected complex value of 167 kDa and suggests that the complex dissociates to some extent during the gel filtration run (Figures 1B and E; Figure S1B). In contrast to individual Msps TOG domains, single TOG1 and TOG2 domains from Stu2p interacted with α/β tubulin heterodimers (Figure 1E; Figures S1D and E). When TOG1 and TOG2 were added in trans in the presence of tubulin, the shift was identical to that produced by a single Stu2p TOG domain with tubulin, indicating that TOG1 and TOG2 were competing for identical or overlapping sites on α/β tubulin (Figure 1E; Figures S1D-H). The Stu2p TOG1-2 construct was insoluble in E. coli and thus precluded further investigation. The co-elution of Stu2p TOG1 with tubulin agrees with findings of Al-Bassam et al., but in contrast to our result, TOG2 did not bind to tubulin in the Al-Bassam study (Al-Bassam et al., 2006).
The EB1 family of plus end binding proteins is characterized by an N-terminal calponin homology (CH) domains, a flexible linker region and a C-terminal dimerization/cargo recruitment domain composed of a coiled coil and four helix bundle (Figure 1A) (Honnappa et al., 2005; Slep et al., 2005). Expressed CH domains (both single and dimerized) from human EB1 and yeast Bim1p did not produce a shift in tubulin elution in the gel filtration assay, suggesting that they do not interact strongly with tubulin (Figures 1C and E; Figures S1I-L). These results agree with other in vitro binding assays with tubulin monomers (Niethammer et al., 2007), although other experiments suggest direct interactions between EB1 and microtubules (Hayashi and Ikura, 2003) and evidence for a microtubule lattice seam interaction has been obtained for the S. pombe homolog Mal3p (Sandblad et al., 2006).
The CLIP-170 family of +TIP proteins is characterized by an N-terminal, conserved Cap-Gly domain (one in yeast and two in higher eukaryotes), a long central coiled-coil, and C-terminal, conserved zinc-finger motifs utilized for cargo attachment (Figure 1A) (Pierre et al., 1992). Functional plus end tracking has been reported for a monomeric construct lacking the dimerization domain, but containing both tandem Cap-Gly domains (Pierre et al., 1994). We found that both a single Cap-Gly domain (CLIP-17057-210) and the tandem Cap-Gly domains from human CLIP-170 (CLIP-1701-350) formed a clear complex with tubulin by gel filtration. However, the distinct elutions for CLIP-17057-210 versus CLIP-1701-350 suggest that tubulin forms a 1:1 and a 2:1 complex with these two constructs respectively. Thus tandem Cap-Gly domains may be capable of multimerizing tubulins (Figures 1D and E, Figures S1M), consistent with previously reported sedimentation behavior of CLIP-170 and tubulin (Arnal et al., 2004; Diamantopoulos et al., 1999).
In summary, the gel filtration tubulin binding studies indicate that TOG and Cap-Gly domains interact directly with α/β tubulin but with varying affinities dependent upon the specific species and the number of tubulin binding domains arrayed or homodimerized. EB1 and Bim1p, in contrast, have sufficiently low affinity for α/β tubulin heterodimers so as to preclude measurement by gel filtration binding studies. However, these results do not rule out a direct interaction of these domains with tubulin in solution or in the microtubule.
Microtubule Nucleation Activity
To investigate functional roles of the +TIP tubulin binding domains, we examined their effects on tubulin polymerization in vitro using turbidity and microscopy as readouts of microtubule formation. We compared and contrasted the effect of single +TIP domains versus tandem domains or artificially homodimerized (fusion to Glutatione S-Transferase (GST) or the GCN4 leucine zipper (LZ) motif) domains.
Msps TOG1-2, the minimum domain that binds to tubulin by gel filtration assays, failed to promote microtubule nucleation (Figure 2A). In contrast, GST-Msps TOG1-2 and an arrayed construct, Msps TOG1-2-1-2, potently promoted microtubule nucleation, greatly reducing the minimal lag time and increasing polymer mass (Figure 2A and D; note, TOG1-2-3-4 could not be expressed in bacteria). Single Stu2p TOG domains failed to promote microtubule polymerization, and the addition of Stu2p TOG1 was even slightly inhibitory (Figure 2A). EB1 CH domain constructs including native homodimer (EB1FL), monomeric (EB11-133) and a truncated homodimer (EB112-255, which lacks a potential autoinhibitory tail sequence and a non-conserved N-terminal segment) failed to affect nucleation rates (Figure 2B). Similar behavior was noted with monomeric Bim1p (Bim1p1-187); however the homodimerized counterpart (Bim1p1-187-LZ) potently promoted microtubule nucleation (Figures 2B and D). Analysis of CLIP-170 constructs showed that a single Cap-Gly domain (CLIP-17057-210) caused slight inhibition of microtubule nucleation, but the homodimerized counterpart (GST-CLIP-17057-210) or a construct containing tandem Cap-Gly domains 1 and 2 (CLIP-1701-350) promoted microtubule formation (similar to findings by Arnal et al. (Arnal et al., 2004) using CLIP-1701-481) albeit with a lag time similar to that observed for tubulin alone (Figures 2C and D). Microscopic analysis (Figure 2D) reveals that +TIPs, particularly CLIP-170, also induce microtubule bundling in vitro, an effect we note will augment the bulk turbidity readings in Figures A-C beyond a comparable concentration of non-bundled microtubules. In summary, our results show that a single +TIP domain has no effect or an inhibitory effect on microtubule nucleation/polymerization, while multimerized domains strongly promote polymerization and often nucleation, with human EB1 being the only exception. Multimerized +TIP domains that promoted microtubule polymerization also promoted polymerization at tubulin’s in vitro critical concentration (data not shown).
Figure 2. +TIP tubulin binding domains promote microtubule nucleation.
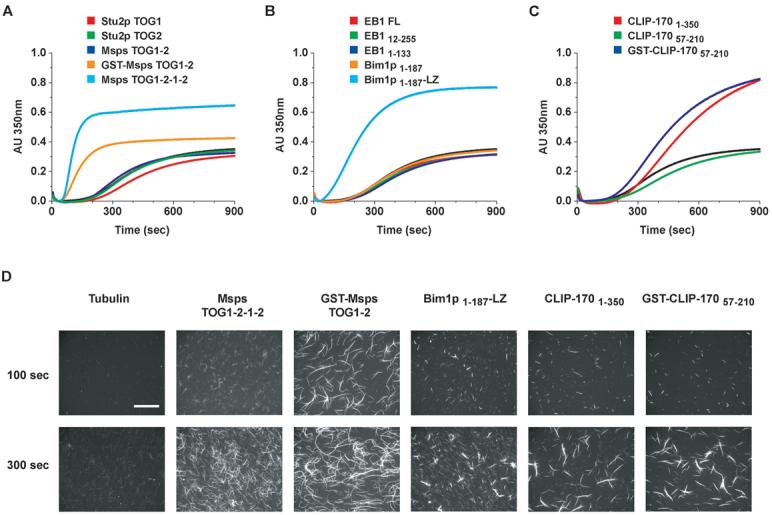
(A-C) Polymerization of tubulin determined via turbidity (absorbance (AU) measured at 350 nm). Tubulin alone (12.5 μM), black curve, tubulin (12.5 μM) plus +TIP construct (1 μM) indicated by color-coded legend above the curves. All +TIPs alone showed no change in turbidity over time and a GST control showed no effect on tubulin’s polymerization rate (data not shown). D) Analysis of TMR-labeled tubulin polymerized for 100 or 300 sec as in (A-C), fixed in glutaraldehyde and pelleted onto coverslips for imaging. Scale bar, 50 μm.
Structure Determination of Tubulin Binding Domains of +TIPs
TOG domains from yeast Stu2p and Drosophila Mini spindles
We attempted to crystallize TOG1, TOG2, and TOG1-2 constructs, but only obtained diffraction-quality crystals from the TOG2 domains of yeast Stu2p and Drosophila Msps. The TOG2 domain from Stu2p crystallized in the space group P212121 with one molecule in the asymmetric unit. The structure was determined using multi-wavelength anomalous dispersion (MAD) phasing from Selenomethionine (SeMet)-derivatized protein to a resolution of 1.7 Å. The TOG2 domain from the Drosophila Msps was crystallized in the space group C2221 (one molecule per asymmetric unit); the structure was determined to 2.1 Å resolution also using MAD phasing. Data, phasing and refinement statistics for these and subsequently described structures are presented in Table 1.
Table 1.
Crystallographic data, phasing and refinement statistics
| Structure | Mini spindles TOG2 | Stu2p TOG2 | EB1 CH Domain | Bim1p CH Domain | CLIP-170 CG 1 | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Space group | C2221 | P212121 | P21 | P212121 | P21212 | ||||||||||
| Cell: a,b,c (Å) | 91.4, 113.4, 78.6 | 28.4, 80.6, 97.2 | 29.3, 65.2, 63.1 (β = 96.2°) | 35.8, 102.5, 31.3 | 27.2, 42.5, 51.8 | ||||||||||
| Wavelength (Å) | 0.97963 | 0.97974 | 1.00801 | 0.97959 | 0.97972 | 1.01627 | 1.12714 | 0.9798 | 1.1271 | 0.7999 | 0.97962 | 0.97979 | 1.03320 | 0.97964 | 1.12713 |
| dmin (Å) | 2.10 | 2.10 | 2.10 | 2.10 | 2.10 | 2.10 | 1.70 | 1.50 | 1.50 | 1.25 | 1.90 | 1.90 | 1.90 | 2.00 | 2.00 |
| % Complete | 97.5 (96.5) |
98.6 (96.4) |
98.2 (91.6) |
97.2 (87.4) |
98.1 (87.4) |
96.1 (87.4) |
99.0 (94.6) |
94.1 (68.7) |
84.9 (31.9) |
97.4 (95.7) |
98.6 (98.5) |
99.7 (98.8) |
98.2 (85.9) |
99.7 (100.0) |
99.4 (99.9) |
| I/σ | 24.6 (3.6) |
25.5 (3.7) |
27.7 (4.3) |
25.2 (6.4) |
23.1 (2.4) |
25.4 (6.1) |
25.5 (9.4) |
20.8 (2.6) |
26.3 (5.2) |
14.9 (3.2) |
33.2 (17.9) |
35.6 (16.2) |
39.3 (12.4) |
22.7 (14.9) |
20.9 (15.0) |
| Rsym (%) * | 3.9 (24.0) |
3.8 (22.9) |
3.5 (18.5) |
4.4 (9.2) |
4.3 (8.9) |
3.9 (7.8) |
4.0 (9.7) |
4.9 (31.5) |
3.5 (14.3) |
5.4 (32.1) |
4.1 (6.7) |
3.9 (7.5) |
3.4 (6.8) |
4.8 (9.2) |
4.6 (7.9) |
|
Phasing power† + Friedel / - Friedel |
1.69 (0.55) / 1.92 (0.61) |
1.99 (0.62) / 2.14 (0.67) |
Ref / 0.27 (0.07) |
2.37 (1.30) / 2.68 (1.48) |
2.92 (1.53) / 3.13 (1.65) |
Ref / 0.47 (0.24) |
3.25 (1.36) / 3.38 (1.46) |
Ref / 0.65 (0.28) |
1.25 (1.16) / 1.50 (1.27) |
2.65 (1.62) / 2.85 (1.69) |
Ref / 0.44 (0.20) |
1.28 (0.87) / 1.37 (0.88) |
Ref / 0.24 (0.07) |
||
|
Figure of merit◇ Centrics / Acentrics |
0.78(0.46) / 0.61(0.25) | 0.87(0.74) / 0.79(0.55) | 0.82(0.76) / 0.72(0.41) | 0.89(0.70) / 0.76(0.58) | 0.72(0.65) / 0.44(0.34) | ||||||||||
| Refinement (Å) | 500-2.10 | 500-1.70 | 500-1.25 | 500-1.90 | 500-2.00 | ||||||||||
| R value ‡ | 20.2 (23.5) | 17.3 (18.7) | 17.4 (19.0) | 19.3 (19.2) | 17.3 (16.2) | ||||||||||
| Rfree § | 21.7 (25.8) | 19.6 (22.6) | 19.8 (21.5) | 22.5 (22.8) | 23.3 (21.3) | ||||||||||
| Rmsd bond lengths (Å) | 0.005 | 0.005 | 0.008 | 0.005 | 0.005 | ||||||||||
| Rmsd bond angles (°) | 1.09 | 1.16 | 1.03 | 1.16 | 1.29 | ||||||||||
| Mean B (min / max) (Å2) | 34.0 (15.0 / 87.7) | 17.2 (5.7 / 59.0) | 12.3 (4.3 / 61.6) | 22.1 (3.9 / 56.1) | 11.2 (1.5 / 51.2) | ||||||||||
| No. protein / solvent atoms | 1852 / 161 | 1991 / 314 | 1974 / 304 | 991 / 101 | 572 / 66 | ||||||||||
Values in parentheses are for the highest resolution shells unless otherwise denoted.
where Ii(h) is the integrated intensity of the ith reflection with the Miller Index h and <I(h)> is the average over Friedel and symmetry equivalents.
MAD phasing power is defined as where P(ϕ) is the experimental phase probability distribution. FN are structure factors at the designated reference wavelength and FD are structure factors of + or - Friedel mates at the other designated wavelength. ΔFh is the difference in heavy atom structure factors between two wavelengths.
Figure of merit is the weighted mean of the cosine of the deviation from αbest.
R value = .
Rfree is calculated using a 10% subset of the data that is removed randomly from the original data and excluded from refinement.
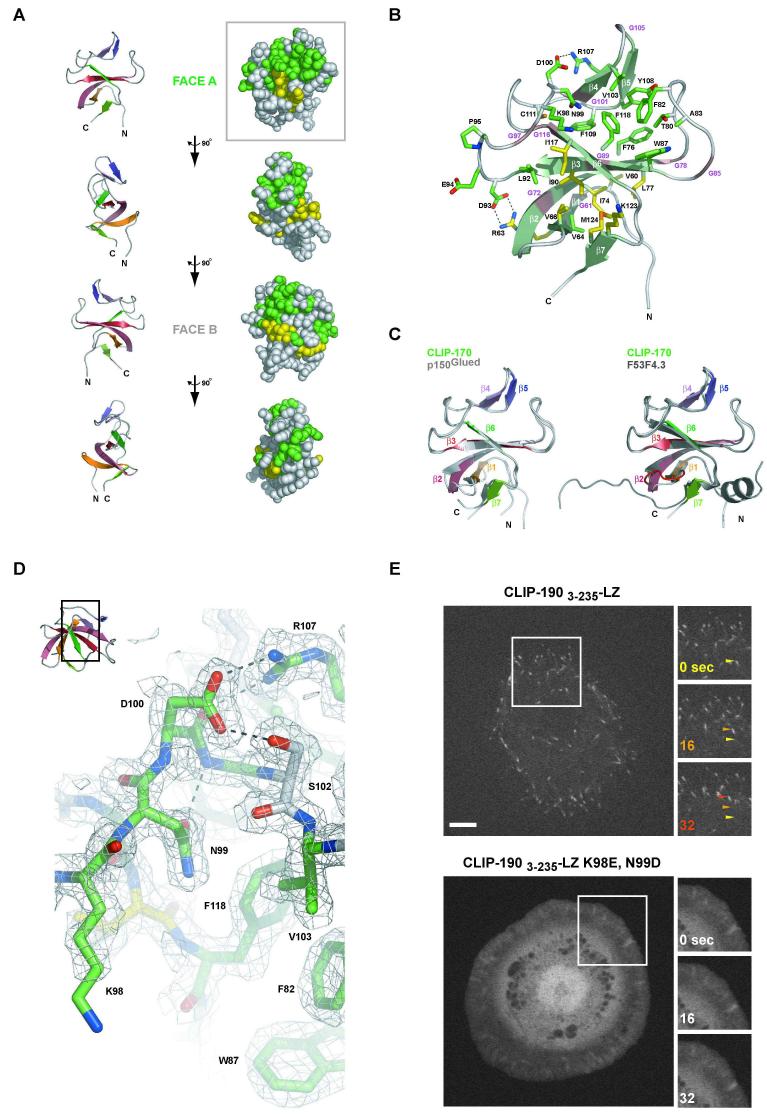
The Stu2p and Msps TOG2 are elongated domains (∼20 × 30 × 60 Å) formed by six HEAT-like repeats A thru F, each comprised of a pair of parallel helices (Figures 3A and B). (We denote the helices using the TOG domain number followed by the HEAT-like repeat letter; fragmented helices are denoted numerically by subscripts and the parallel helix is denoted by a prime.) Only HEAT repeats C and D of Msps TOG2 and C and F of Stu2p form canonical HEAT repeats, in which the first α helix is kinked by 90°, positioning the N-terminal segment of the helix orthogonal to helix α’ (Figure 3D), structurally similar to the third orthogonal helix found in armadillo repeats. HEAT-like repeat E, particularly of Stu2p, is characterized by a segmented N-terminal α helix we denote as α1 and α2 (Figure 3D). The TOG domain has a small helical twist along the axis of the HEAT-like repeats, compared with the more substantial twist of other HEAT-repeat structures such as importin-β (Cingolani et al., 1999).
Figure 3. Structure of the second TOG domains from Mini spindles and Stu2p.
Ribbon diagram of the TOG2 domain from Msps (A) and Stu2p (B) with the six HEAT-like repeats represented in shades of similar color and labeled A-F. The conserved and non-conserved regions (Faces A and B respectively) are indicated. C) Least-squares fit of Msps (color) and Stu2p (grey) with TOG 2 domains shown in cylindrical helix representation. D) Individual TOG2 HEAT-like repeats are shown for Msps and Stu2p in ribbons format in similar orientations after global least squares fit of each TOG2 domain. The definitive α helix kink that defines HEAT repeats is evident in the α2 helices of Msps HEAT-like repeats C and D and Stu2p HEAT-like repeats C and F. E) 90° rotations of the Msps TOG2 domain about its long axis shown at left in ribbons for orientation and at right in CPK representation for conservation mapping. TOG2 residues with 80% identity across species are represented in green, 80% conservation in yellow (see Figure S2). E) 2Fo-Fc electron density map at 1.7 Å resolution of the Stu2p TOG2 structure contoured at 1.0 σ showing the surface exposed and highly conserved KEKK loop of HEAT-like repeat C. G) 2Fo-Fc electron density map at 2.1 Å resolution of the Msps TOG2 structure contoured at 1.0 σ showing the surface exposed W292 residue and the buried R295 - D331 salt bridge. Inset (upper left) indicates the relative orientation of the TOG domain (F and G). H) Gel filtration tubulin binding assays for wild type (WT) and mutant Msps TOG1-2. Single or double mutations of the conserved TOG domain tryptophan (TOG1: W21E, TOG2: W292E) are indicated above the chromatogram. Tubulin alone, black; Msps TOG1-2 WT alone, red. The plot indicates absorption at 280 nm on the y-axis (mAU) and elution volume in ml along the x-axis.
Interesting and likely important features of the TOG domain are the structured loops between the helices. The intra-HEAT loops on one face of the molecule (face A) show a higher degree of structural identity between Stu2p and Msps TOG2 (r.m.s.d. of 1.3 Å over 60 mainchain atoms)(Figure 3C) than the loops on the opposite B face (r.m.s.d. of 3.7 Å over 87 homologous mainchain atoms, not factoring the larger Stu2p inserts that occur in face B) and the helices themselves (r.m.s.d. of 2.1 Å over 513 mainchain atoms). The loops in face A also contain the most highly conserved residues among TOG domains from many species (Figure 3E). Among these are seven highly conserved lysines and an arginine, making the net charge on face A highly positive (Figure 3F, Figure S2). Also exposed on face A is an invariant tryptophan residue, W292, found in TOG types A and B (type C has a conserved phenylalanine), situated on the intra-HEAT loop between α2A2 and α2A’ (Figure 3G, Figure S2). W292 along with valine V334 establishes a conserved hydrophobic character to face A. Directly below W292, a highly conserved, buried salt bridge between R295 and D331 prohibits the tryptophan from torsional engagement with the core (Figure 3G).
The combination of highly conserved, exposed hydrophobics and positively charged residues makes face A a likely region for interacting with tubulin. We tested this hypothesis through the single or double mutagenesis of the conserved tryptophans exposed on face A of Msps TOG1 and TOG2 (W21 and W292 respectively) to glutamates. The ability of Msps TOG1-2 to shift α/β tubulin over gel filtration was diminished with either single point mutant (W21E and W292E) and completely abrogated in the double point mutant (Figure 3H). This result indicates that the solvent-exposed conserved tryptophan is a critical determinant for tubulin binding on face A and that binding is a cooperative activity between tubulin heterodimers and the arrayed TOG domains. While this paper was being submitted, Al-Bassam et al. reported the structure of the TOG3 domain from Zyg-9, the XMAP215 homolog in C. elegans. This class B TOG domain displays an overall fold and tubulin binding activity that corroborates our Msps and Stu2p TOG2 domain results (Al-Bassam et al., 2007).
CH Domains of Yeast Bim1p and Human EB1
The CH domains of EB1 and Bim1p structures were determined using MAD phasing and SeMet derivatized protein. The structure of human EB1 is very similar to a previously reported structure (Hayashi and Ikura, 2003), but ours was determined with two molecules in the asymmetric unit and to a higher resolution of 1.25 Å. We also delineate two helices, a 310 helix (between α3 and α4) and α7, which were modeled as loop regions in the Hayashi et al. structure. Bim1p was determined from a P21212 lattice to a resolution of 1.9 Å with one molecule in the asymmetric unit. The overall fold of the Bim1p CH domain is nearly identical to the human with an r.m.s.d. of 1.3 Å over 351 mainchain atoms (Figure 4C, compared using EB1 protomer A) while the comparable r.m.s.d. between our human EB1 and the Hayashi et al. structure is 0.5 Å. The EB1/Bim1p CH domain is formed by eight helices that pack around a central conserved hydrophobic helix, α3. Helices 3,4 and 6 are aligned in parallel, with α4 flanked on one side by an extended loop dissected by the 310 helix and flanked on the other by the extended α4-5 loop (Figures 4A and B).
Figure 4. Structure of the calponin homology domains of EB1 and Bim1p.

Ribbon diagram of the N-terminal CH domain from EB1 (A) and Bim1p (B) centered on the highly conserved α6 helix. C) Least-squares fit of EB1 (color) and Bim1p (grey) CH domains with cylindrical helix representation showing the overall structural conservation between these two members. D) 2Fo-Fc electron density map at 1.25 Å resolution of the EB1 CH domain structure contoured at 1.0 σ showing the highly conserved aromatic core. E) 90° rotation of the EB1 CH domain about the y-axis shown above in ribbon format for orientation and below in CPK representation for conservation mapping and to summarize mutagenesis results. Center: CH domain residues with 80% identity across species are represented in green, 80% conservation in yellow (D-E)(see Figure S3). Bottom row: results of CH domain mutagenesis on the ability of EB11-187-LZ to plus end track are mapped: ablation of microtubule association: brick red (three cluster mutants indicated: [S16A, R17E, H18E, D19R], [K59E, K60E], [K66E, L67D]); no effect on microtubule plus end tracking: slate.
Many of the highly conserved surface residues in EB1 proteins are found in α6, featuring a conserved hydrophobic groove created by Phe107, Trp110 and Phe114 (Figure 4D) and conserved electrostatic residues Gln102, Asp103, Glu106, Gln109 and Lys113 (not shown). To explore the tubulin binding interface of EB1, we mutated several of the conserved surface residues using single or cluster mutations, transfected these EGFP-tagged constructs into HeLa cells, and examined plus end tracking by time lapse microscopy. To prevent heterodimerization with native EB1, the endogenous dimerization domain was replaced with the GCN4 leucine zipper motif. Mutations on one hemisphere of the domain (delineated as face A) resulted in ablation of in vivo plus end tracking activity (mapped in Figure 4E), yielding instead, diffuse cytosolic localization. This region is delineated by α1 and the α3-α4 loop that encompasses the 310 helix described earlier. This tubulin binding zone partially overlaps with the CH domain’s actin binding zone delineated by the regions homologous to α1 and α5-α6 in EB1 (Figure S4)(Mino et al., 1998; Sutherland-Smith et al., 2003). We note that the majority of EB1’s surface is highly conserved, likely reflecting its use in a variety of conserved protein-protein interactions in addition to tubulin binding (Ligon et al., 2006; Vaughan, 2005).
Cap-Gly Domain of Human CLIP-170
The first Cap-Gly domain from human CLIP-170 was crystallized in the space group P21212 with one molecule in the asymmetric unit, and the structure was determined to 2.0 Å resolution by MAD phasing (using a SeMet-derivatized mutant, Q124M). The fold, which has a weak similarity to a SH3 domain, is composed of seven β-strands that form two β-sheets (Figures 5A and B). Strands are connected by long, extended loop regions with conserved glycine residues mediating key structural turns. The central β-sheet bifurcates the conserved hydrophobic core and undergoes a sharp bend in β2 enabling the β2-1-7 region of the β-sheet to envelop the lower half of the conserved hydrophobic core. The overall architecture is highly similar to the structure of the p150Glued Cap-Gly domain and a Cap-Gly domain from a putative C. elegans α-tubulin folding chaperone, F53F4 (respective r.m.s.d. values of 0.9 Å and 1.1 Å over 213 mainchain atoms; Figure 5C)(Hayashi et al., 2005; Li et al., 2002).
Figure 5. Structure of the first Cap-Gly domain of CLIP-170.
A) 90° rotation of CLIP-170′s first Cap-Gly domain about the y-axis shown above in ribbon format for orientation (left) and in CPK representation for conservation mapping (right) with 80% identity shown in green, 80% conservation in yellow (see Figure S4). B) Ribbon diagram of the first Cap-Gly domain from CLIP-170 with β-strands indicated in light green. Conserved glycine residues that constitute the Cap-Gly domain are indicated along the ribbon in magenta. Conserved residues across Cap-Gly domains (excluding glycines) are indicated in stick format (colored as in A). The D100-R107 and R63-D93 salt bridges are indicated. C) Least squares fit of the CLIP-170 Cap-Gly 1 domain (colored) and the human p150Glued Cap-Gly domain (grey, left)(Honnappa et al., 2006) and the C. elegans F53F4.3 Cap-Gly domain (grey, right) (Li et al., 2002). The loop insert present in F53F4.3 is indicated in red. D) 2Fo-Fc electron density map at 2.0 Å resolution, contoured at 1.0 σ showing a segment of the conserved β3-β4 loop’s GKNDG motif with D100 torsionally fixed via hydrogen bonds to R107 and S102 and the N99 rotamer fixed via a hydrogen bond between its δO and G101’s backbone amine. The GKNDG motif flanks the conserved hydrophobic region F82, W87, V103 and F118 as shown, that together comprise the tentative tubulin C-terminal binding site. Inset (upper left) indicates the relative orientation of the Cap-Gly domain. E) Images of Drosophila S2 cells transfected with CLIP-1903-235-LZ-EGFP (above) and the K98E, N99D mutant (below). Boxed regions at left are shown as time series at right (time in sec. indicated in the upper left). Arrows track a single microtubule tip for the CLIP-1903-235-LZ-EGFP construct across the time series, color-coded according to the time point. Scale bar, 5 μm.
While both halves of the hydrophobic core are highly conserved across Cap-Gly domains, the upper half’s constituency is conserved to a higher degree with core aromatic residues including Phe76, Phe82, Trp87, Tyr108, Phe109 and Phe118 and structural elements including the β4-β5 sheet and the extended β2-β3 loop. Comparing Cap-Gly structures using only the highly conserved segment from β2 to β6 (CLIP-17072-118), CLIP-170 exhibited an r.m.s.d. of only 0.4 Å with both p150Glued and F53F4.3.
The most highly conserved face of the Cap-Gly domain consists of conserved, exposed hydrophobic residues of the upper core and an invariant region in the β3-β4 loop, containing the sequence GKNDG (residues 97-101)(Figure 6D). The GKNDG motif flanks the exposed hydrophobic core and is stabilized by two conserved salt bridge pairs: Arg63-Asp93 and Asp100-Arg107 (Figures 5A, B and D). A co-crystal of the p150Glued Cap-Gly domain with the C-terminal dimerization domain of EB1 has revealed that the GKNDG Cap-Gly sequence interacts with the sequence motif EEY/F-COO- on EB1 (Honnappa et al., 2006). This EEY/F-COO- motif is also found in α-tubulin, and thus it was proposed that the same interface occurs between Cap-Gly domains and tubulin. To test if this conserved GKNDG motif contributed to tubulin binding and plus-end localization, we altered the charge of this segment by mutating the motif to GEDDG. Using the plus end tracking competent Drosophila construct CLIP-1903-235-LZ-GFP, the GEDDG mutant failed to plus end track or localize to microtubules but was instead diffusely localized in the cytoplasm (Figure 6E; Movie S1). This result substantiates the hypothesis of Honnappa et al. for the role of this motif in microtubule interactions and suggests that EB1 and tubulin compete for the same binding site.
Figure 6. In vivo analysis of +TIP tubulin binding domains and the plus end tracking correlate.
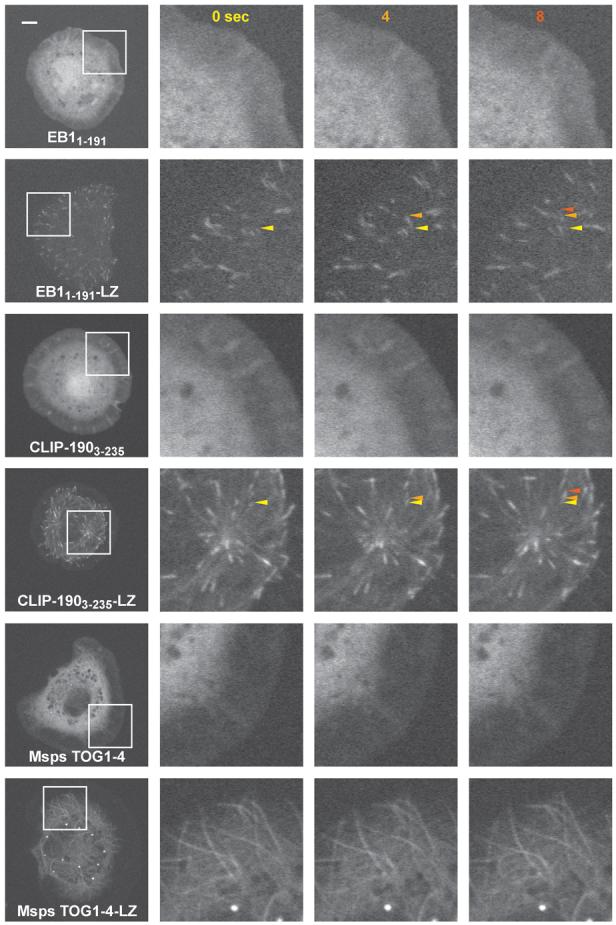
In vivo analysis of Drosophila S2 cells transfected with EGFP fusions of +TIP domains or +TIP domains fused to the GCN4 leucine zipper (LZ) motif. Domains from top to bottom: DmEB11-191, CLIP-1903-235 and Msps TOG1-4 (residues 3-1175). Images at right correspond to a magnification of the boxed region in the first column and represent a time series at 4 sec intervals denoted at the top of each column. Arrows, color-coded to the respective time point, track individual microtubule plus ends. Scale bar in left column, 5 μm.
Multimerized +TIP Domains Confer Plus End Tracking In Vivo
Having determined structures and in vitro activities of +TIP domains, we next tested the functions of these domains in living cells with regard to plus end tracking activity. A single CH domain from EB1 or Cap-Gly domain from CLIP-190 fused to GFP did not plus end track when transfected into Drosophila S2 cells (Figure 6). However, artificial dimerization of these domains with a leucine zipper enabled clear plus end tracking along microtubules (Figure 6; Movies S2 and S3). We also tested the ability of the TOG domains from Msps to confer plus end localization in S2 cells. Consistent with previous findings (Popov et al., 2001), we found that full length Msps fused to GFP could plus end track, but a truncation encompassing TOG1-4 did not and instead was diffusely localized in the cell (Figure 6) as was a leucine zipper dimerized TOG1-2 construct (not shown). This result, unlike the above result for EB1 and CLIP-170, suggests a more complex plus end tracking requirement for XMAP215 than simply binding multiple tubulin monomers and suggests a possible synergistic role for the C-terminal domain (CTD) to cooperate with the TOG domains for microtubule plus end localization. Analysis of Msps TOG1-4 fused to a leucine zipper however, resulted in microtubule decoration. This construct raised the functional number of TOG domains to eight indicative that TOG domains have an inherent affinity for the microtubule lattice, but the bona-fide microtubule bind and release mechanism employed by XMAP215 family +TIPs likely requires a carefully titrated affinity mediated by five TOG domains and the CTD in metazoans and four TOG domains and two C-terminal tubulin binding domains in the functionally homodimeric yeast counterparts.
To obtain further evidence for a role of dimerization in plus end tracking, we fused the human EB1 CH domain to either FRB or FKBP domains, transfected both constructs into HeLa cells and then induced dimerization in real time by addition of the natural product rapamycin (which binds to and crossbridges FRB and FKBP (Banaszynski et al., 2005; Choi et al., 1996)). When either construct alone was transfected into HeLa cells, only diffuse localization was observed either in the presence or absence of rapamycin (data not shown). When both constructs were transfected simultaneously in the absence of rapamycin, a diffuse cytoplasmic localization was again observed. However, when rapamycin was added, the GFP-tagged proteins rapidly relocalized to the tips of growing microtubules (Figure 7; Movies S5A and B). Identical results were obtained with a similar set of experiments chemically dimerizing Cap-Gly domain 1 of CLIP-170 (CLIP-1703-210-FRB) to Cap-Gly domain 2 (CLIP-170129-350-FKBP) in HeLa cells (Figure 6B; Movie S6). These results provide strong evidence for the role of dimerization of +TIP tubulin binding domains in the localization to microtubule plus ends.
Figure 7. In vivo chemical dimerization facilitates microtubule plus end tracking in real time.
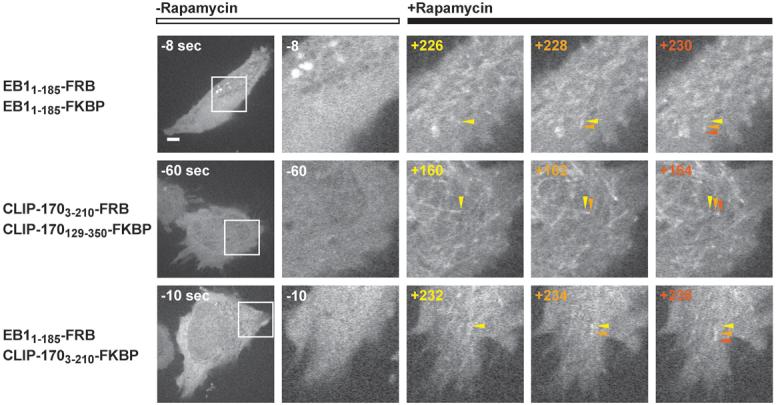
HeLa cells dually transfected with FRB-EGFP and FKBP-EGFP fusions of EB1 and/or CLIP-170. EB1 constructs embody residues 1-185, CLIP-170 constructs residues 3-210 and 129-350. After one minute of imaging, rapamycin was added to the media for a final concentration of 50 nM. Magnified images at right correspond to the boxed region in the first column of the respective row. Images represent a single time point pre-rapamycin treatment and three consecutive time points, taken at 2 sec intervals, post-rapamycin treatment. Arrows, color-coded to the respective time point, track individual microtubule plus ends. Time in sec is indicated relative to rapamycin addition (t = 0 sec). Scale bar in left column, 5 μm.
Given the similar minimal requirement for two tubulin binding domains for plus end tracking by EB1 and CLIP-170, we next asked whether a heterodimer of these two unrelated structural domains might enable plus end tracking. To address this, we co-transfected EB11-187-FRB and CLIP-1703-210-FKBP into cells. Remarkably, rapamycin addition also induced a relocalization of these proteins to microtubule tips (Figure 7, Movie S7). These results indicate that two structurally unrelated +TIP tubulin binding scaffolds can cooperate together to localize to growing microtubule plus ends.
DISCUSSION
In this work, we have analyzed the biochemical mechanisms of three plus end tracking protein families (EB1, CLIP-170 and XMAP215) using x-ray crystallography, in vitro tubulin binding/assembly assays, and in vivo plus end tracking assays. We present five crystal structures of +TIP tubulin binding domains across multiple species and families illustrating structural conservation within families and functional convergence across families. In vitro and in vivo analysis reveal an underlying general theme to these unrelated domains. While tubulin binding can be conferred by a single +TIP domain, the promotion of microtubule nucleation and polymerization in vitro and plus end tracking in vivo require multiple domains acting in concert. In the case of EB1 and CLIP-170 members, two domains are required for robust plus end localization and these two structurally unrelated domains can even cooperate if artificially joined together to achieve this activity. XMAP215 members are more complicated, perhaps due to the relative weaker association between TOG domains and tubulin in higher eukaryotic members. However, multimerization of TOG domains clearly facilitates tubulin nucleation in vitro and overall microtubule association in living cells. From these data, we propose a model in which unrelated tubulin binding domains have convergently evolved a similar mechanism for enhancing tubulin assembly on growing microtubule ends by acting as multivalent tubulin polymerization chaperones.
Structural comparison of +TIP proteins
The tubulin binding domains of these three +TIP proteins are quite diverse in their architecture and show no evidence of a common evolutionary origin. However, we show that the structural conservation within a +TIP class is very high, as evidenced by the near superimposition of yeast Bim1p with human EB1 and yeast Stu2p with Drosophila Mini spindles. The different architectures also suggest varying interactions modes with tubulin and potential synergy between the +TIPs. XMAP215 has a very flat binding interface created by a series of rigid, highly conserved loops on one face of the TOG domain; the length of the TOG domain is fairly closely matched to a tubulin monomer (α or β) and likely makes extensive contacts through a combination of hydrophobic and electrostatic interactions. In contrast to the elongated TOG domain, the CH domain of EB1 is spherical and approximately half the size. Mutagenesis suggests that several residues around one hemisphere contribute to tubulin binding. This data is consistent with recent work suggesting that EB1 might nestle in the groove between tubulin protofilaments (Sandblad et al., 2006). CLIP-170 also has a globular structure and our data, in conjunction with others (Honnappa et al., 2006; Peris et al., 2006), suggests that it might bind tubulin’s disordered C-terminal tail as part of its binding interface.
Comparison of the +TIPS and our protein engineering studies suggest a considerable structural variation in how +TIP proteins can be interconnected to achieve their activities in plus end tracking and promoting tubulin assembly. In some cases, tubulin binding domains are arrayed in tandem along a polypeptide chain with presumably unstructured linkers in between (e.g. Mini spindles, Ch-TOG, CLIP-170). The cis linking domains within a polypeptide appears to be critical, since we show that TOG domains added in trans do not reconstitute function. In other cases, single α/β tubulin binding domains are found in a polypeptide and multivalent tubulin binding is achieved by polypeptide dimerization (e.g. EB1 family and Bik1 (the CLIP-170 homologue from yeast)). Hybrid strategies are also employed (e.g. CLIP-170, which has tandem Cap-Gly domains and is dimerized via a coiled coil and Stu2p which has tandem TOG domains, a dimerization domain and an additional C-terminal microtubule binding domain). We also find that native dimerization sequences are not essential, as a variety of artificial dimerization strategies (e.g. GCN4 leucine zippers, gluthione S-transferase, and FKBP-rapamycin-FRB) are capable of reconstituting +TIP protein function.
Collectively, these studies reveal structural variation in how multiple +TIP tubulin binding domains can be combined to promote tubulin oligomerization for microtubule assembly. However, cis versus trans arrangements of +TIP domains may produce certain unique outcomes. For example, the linear arrays of TOG domains in the extended XMAP215 structure (Cassimeris et al., 2001) may generate a pseudo protofilament-like arrangement that is particularly effective for microtubule nucleation. Indeed, we have found XMAP215 has a potent microtubule nucleation ability compared with CLIP-170, potentially related to an in vivo nucleation activity given XMAP215’s TACC-dependent localization to the centrosome (Lee et al., 2001). Analogous mechanisms exist for the actin cytoskeleton where the Spire protein utilizes several arrayed actin binding domains to template the nucleation of an actin filament (Quinlan et al., 2005).
+TIP proteins as microtubule polymerization chaperones
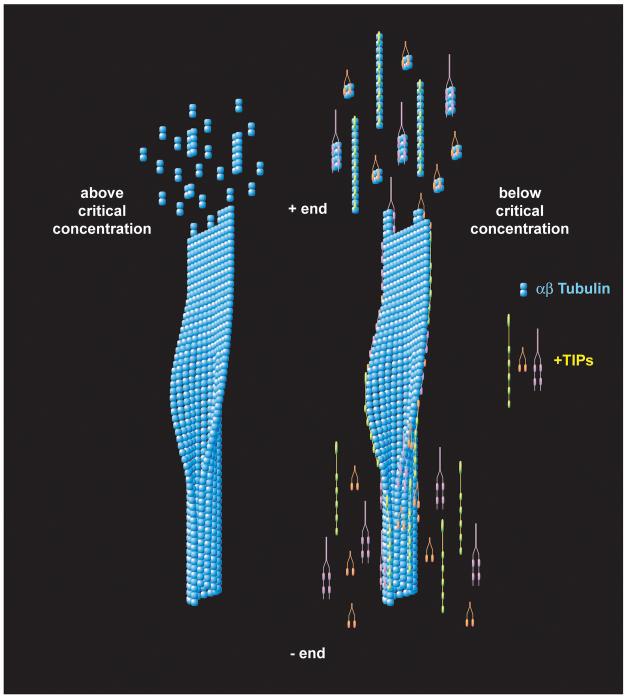
Our data show that single +TIP tubulin binding domains do not promote microtubule nucleation or growth in vitro (and in some cases are inhibitory), nor do they localize to microtubule plus ends in vivo. In contrast, multimerized +TIP tubulin binding domains are potent microtubule nucleator in vitro, promote microtubule growth in vitro and are requisite in vivo for microtubule plus end localization. For EB1 and CLIP-170, we dynamically show that plus end tracking requires an ability to bind more than one tubulin subunit. These results suggest a model (Figure 8) in which +TIPs bind multiple tubulin dimers in solution and then deliver these larger tubulin oligomers to the ends of microtubules. This general idea was first introduced two decades ago by Gard and Kirschner (Gard and Kirschner, 1987) to explain the high rates of tubulin assembly induced by XMAP215. Multimerization overcomes the inherent polymerization barrier tubulin heterodimers face due to single longitudinal and lateral tubulin:tubulin affinities estimated to be on the order of mM and M respectively (Sept et al., 2003; VanBuren et al., 2002). +TIP-induced multimerization of tubulin would increase the effective affinity for the microtubule lattice through cooperative binding, thereby decreasing the critical concentration for polymerization. By stabilizing tubulin-tubulin interactions prior to their full incorporation into a mature, cylindrical lattice, the +TIPs would act as polymerization chaperones. Physiologically, such chaperones would become particularly important for enhancing the growth of microtubules when free tubulin dimers are below the critical concentration of microtubule assembly or when microtubule destabilizing proteins are active (e.g in mitosis (Rogers et al., 2002; Tirnauer et al., 2004; Tournebize et al., 2000)). This mechanism also enhances the spatial and temporal regulation of microtubule assembly in the cell, in part by regulating the localization and/or activity of +TIPs without modifying tubulin, the basic unit of polymerization.
Figure 8. Dimerization model for plus end tracking and chaperoned tubulin polymerization.
Model for +TIP chaperoned tubulin polymerization. Above tubulin’s critical concentration, αβ tubulin heterodimers preferentially bind the microtubule plus end resulting in microtubule polymerization and growth. Below tubulin’s critical concentration, +TIP-mediated multimerization of αβ tubulin effectively raises the affinity for the microtubule plus end enabling multimer subunit addition to the microtubule plus end. Chaperone-mediated co-polymerization of +TIPs facilitates localization of +TIPs at the microtubule plus end. A lower affinity for the microtubule than for free multimerized tubulin subunits leads to +TIP dissociation and the apparent plus end tracking behavior noted for fluorescent-tagged +TIPs. XMAP215, EB1 and CLIP-170 family members are modeled as tethered tubulin binding domains (colored yellow, orange and purple respectively). A diverse array of oligomeric tubulin binding strategies (lateral vs. longitudinal) is likely for each +TIP family given the tethered nature of their domains. For simplicity, a single oligomeric tubulin interaction is modeled for each +TIP family. See Discussion for details.
In addition to our work, support for this general model comes from a number of other laboratories. Work by Perez et al. first showed that tandem Cap-Gly domains in CLIP-170 plus end track (Perez et al., 1999) whereas single Cap-Gly domains of the homolog CLIP-115 show greatly reduced microtubule association (Hoogenraad et al., 2000). More recently, optical trapping studies by Kerssemakers et al. analyzing microtubule growth (Kerssemakers et al., 2006) showed step-wise growth that increased in size in the presence of full length XMAP215, a result they interpreted as XMAP215-facilitated incorporation of tubulin oligomers onto the microtubule end.
An apparent exception to the multiple tubulin binding rule for +TIPs is the CLASP family, which contains only one TOG domain at its N-terminus (Akhmanova et al., 2001). However, informed by our TOG structures and secondary structure predictions we can identify several TOG-like (TOGL) domains in the CLASP family. Two additional dodeca-helical domains are predicted with alternating loops that exhibit high homology to TOG domain intra-HEAT loops (see Figure S6 and legend). If folded into a HEAT-like structure, the conservation profile of alternating loops would be localized to one face of the domain, suggesting that TOGL domains may bind tubulin by a mechanism similar to what we describe here for the TOG domain. Thus, although poorly conserved at the primary structure level, XMAP215 and CLASP appear to be ancient +TIP relatives that may employ a similar general strategy for plus end tracking. Support for CLASP’s possible polymerization chaperone role comes from Maiato et al. who observed CLASP-dependent microtubule subunit incorporation into fluxing kinetochore fibres (Maiato et al., 2005).
Our model for +TIPs as multimeric tubulin chaperones undoubtedly oversimplifies the complexity of interactions that are occurring at the microtubule plus end in vivo. +TIP families show divergent effects on microtubule dynamics, some promoting growth while others act as anti-pause, destabilization, rescue and perhaps even nucleation factors. Some of these differences in activity may reside in variations in the affinity constants of +TIP domains with tubulin monomers, oligomers and microtubules. As an example, non-EB1 family microtubule-binding CH domain proteins, including CLAMP and HEC1 appear to have higher affinity for the microtubule lattice than EB1 and localize along the entire length of the microtubule rather than just at plus ends (Dougherty et al., 2005; Wei et al., 2007). The spatial arrangement of +TIP domains, arrayed versus dimerized, may lead to distinct effects on microtubule dynamics by preferentially stabilizing lateral versus longitudinal tubulin-tubulin interactions. Finally, the intricate web of +TIP-+TIP interactions (Wolyniak et al., 2006) may generate unique outcomes on tubulin assembly. Multi-component in vitro assays, high resolution EM analysis of microtubules, and x-ray structures of +TIP-tubulin complexes will be required to further understand how +TIPs regulate the microtubule lattice.
EXPERIMENTAL PROCEDURES
Constructs, Crystallization and Structure Determination
Protein constructs, expression, crystallization, x-ray data collection, and methods of solving the structures are described in Supplementary Methods.
Gel Filtration Binding Assays
Tubulin binding assays were conducted over gel filtration using a Tricorn Superdex 200 10/30 column (GE Healthcare) equilibrated in running buffer (20 mM Pipes pH 6.5, 0.1 % β-ME, 200 mM KCl, 2 mM MgCl2, 1 mM EGTA, 50 μM GTP) and maintained at 4° C. 200 μl containing +TIP protein (10 μM) and tubulin (5 μM) were incubated for 20 min at 4° C in running buffer with 150 μM GTP (Bim1p1-187 and Bim1p1-187-LZ were also supplemented with 100 mM KCl to improve solubility) and injected onto the gel filtration column with a 0.3 ml/min flow rate. Gel filtration assays coupled with in line dynamic light scattering were performed using a Shodex KW-803 gel filtration column coupled to a Dawn EOS (Wyatt Technologies) run at 0.5 ml/min in running buffer.
Microtubule Polymerization Assays
Turbidity experiments were performed with 12.5 μM tubulin and 25 μM of the indicated +TIP proteins. Purified proteins were incubated at 4° C in assembly buffer (50 mM MES, pH 6.6, 3.4 M glycerol, 5 mM DTT, 1 mM EGTA, 5 mM MgSO4, 1 mM GTP) for 15 min and then injected into 37° C quartz cuvettes. Absorbance at 350 nm was monitored at 0.7 sec intervals. Initial A350 spikes resulted from sample injection and were taken as t=0. A350 returned to baseline within 20 sec and the 30 sec point was utilized for baseline adjustment.
Microscopy-based analysis of microtubule polymerization used identical protein concentrations as the turbidity experiments; however, 10% of the final tubulin concentration was rhodamine-labeled (Cytoskeleton, Inc.). Reactions were incubated at 37° C in assembly buffer and aliquots were removed at t=100 and 300 sec and diluted 11-fold into assembly buffer plus 1% glutaraldehyde at 37° C for 3 min. Fixation was quenched by addition of Tris pH 7.0 to a final concentration of 100 mM and subsequently diluted 11-fold into glycerol-free assembly buffer. Microtubules were adhered onto poly-lysine coated coverslips by layering 20 μl of reaction mixture above a 20% glycerol cushion (in assembly buffer) and centrifuging at 24,000 × g for 30 min. Coverslips were subsequently fixed with -20° C methanol, mounted and imaged using an inverted Zeiss Axiovert 200M microscope, 40x 1.3 N.A. oil objective, SensiCam camera (Cooke) and μManager software (www.micro-manager.org).
Cell Culture, Transfection, and Live Cell Imaging
HeLa cells and Drosophila S2 cells were grown in Dulbecco’s modified Eagle’s solution and Schneider’s Drosophila medium respectively, supplemented with 10% fetal bovine serum. HeLa and S2 cells were transfected using Lipofectamine (Invitrogen) and Cellfectin (Invitrogen) respectively according to manufacturer’s protocols. For live cell imaging, cells were plated on Matek dishes (pre-coated with ConA for S2 imaging) and images were acquired using a Zeiss Axiovert microscope outfitted with a spinning disk scanhead (Yokogawa) and 488 nm argon laser (Solamere Technologies), a Hammamatsu Orca ERG2 camera or a Mega10 intensified CCD camera (Stanford Photonics), 100x 1.45 N.A. oil objective (maintained at 37° C by an objective heater (Bioptechs) for HeLa imaging) and Metamorph software (Molecular Devices) using 2x binning for imaging EGFP fusion proteins. Movies were taken at frame intervals of either 2 or 3 sec and processed using ImageJ and Photoshop (Adobe). For rapamycin-mediated heterodimerization experiments, DMSO or ethanol-solubilized rapamycin (Calbiochem) was added to the media to yield a final concentration of 50 nM. Previously reported inhibition of CLIP-170 microtubule association via rapamycin treatment was not evident for our CLIP-170 FRB and FKBP fusion constructs, likely due to the robust nature of the chemical dimerization assay or absence of specific rapamycin-sensitive phosphorylation sites in these constructs (Choi et al., 2002).
Structure coordinates for Msps TOG2, Stu2p TOG2, EB1 CH, Bim1p CH and CLIP-170 CG-1 domains have been deposited in the Protein Data Bank under accession codes 2QK2, 2QK1, 2QJZ, 2QJX and 2QK0 respectively.
Supplementary Material
ACKNOWLEDGMENTS
This work was facilitated by use of the Lawrence Berkeley National Laboratory Advanced Light Source beamlines 8.3.1 and 8.2.2 and the generous support of the 8.3.1 staff: James Holton, George Meigs and Jane Tanamachi. Amino acid sequencing of the plasmin-cleaved Msps construct was performed by Nancy Williams at the W.M. Keck Foundation Biotechnology Resource Lab at Yale University. The authors thank Nicole Mahoney for stimulating discussions. This work has been supported by the National Institutes of Health (NIH38499 [R.D.V.]), the Agouron Institute (K.C.S.) and the Helen Hay Whitney Foundation (K.C.S.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DEDICATION This paper is dedicated to the memory of Thomas Kreis, whose initial discovery of the CLIP proteins and their localization to the plus end launched the field of +TIP proteins.
REFERENCES
- Akhmanova A, Hoogenraad CC, Drabek K, Stepanova T, Dortland B, Verkerk T, Vermeulen W, Burgering BM, De Zeeuw CI, Grosveld F, Galjart N. Clasps are CLIP-115 and -170 associating proteins involved in the regional regulation of microtubule dynamics in motile fibroblasts. Cell. 2001;104:923–935. doi: 10.1016/s0092-8674(01)00288-4. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, Larsen NA, Hyman AA, Harrison SC. Crystal Structure of a TOG Domain: Conserved Features of XMAP215/Dis1-Family TOG Domains and Implications for Tubulin Binding. Structure. 2007;15:355–362. doi: 10.1016/j.str.2007.01.012. [DOI] [PubMed] [Google Scholar]
- Al-Bassam J, van Breugel M, Harrison SC, Hyman A. Stu2p binds tubulin and undergoes an open-to-closed conformational change. J Cell Biol. 2006;172:1009–1022. doi: 10.1083/jcb.200511010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andrade MA, Petosa C, O’Donoghue SI, Muller CW, Bork P. Comparison of ARM and HEAT protein repeats. J Mol Biol. 2001;309:1–18. doi: 10.1006/jmbi.2001.4624. [DOI] [PubMed] [Google Scholar]
- Arnal I, Heichette C, Diamantopoulos GS, Chretien D. CLIP-170/tubulin-curved oligomers coassemble at microtubule ends and promote rescues. Curr Biol. 2004;14:2086–2095. doi: 10.1016/j.cub.2004.11.055. [DOI] [PubMed] [Google Scholar]
- Banaszynski LA, Liu CW, Wandless TJ. Characterization of the FKBP.rapamycin.FRB ternary complex. J Am Chem Soc. 2005;127:4715–4721. doi: 10.1021/ja043277y. [DOI] [PubMed] [Google Scholar]
- Brittle AL, Ohkura H. Mini spindles, the XMAP215 homologue, suppresses pausing of interphase microtubules in Drosophila. Embo J. 2005;24:1387–1396. doi: 10.1038/sj.emboj.7600629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch KE, Hayles J, Nurse P, Brunner D. Tea2p kinesin is involved in spatial microtubule organization by transporting tip1p on microtubules. Dev Cell. 2004;6:831–843. doi: 10.1016/j.devcel.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Carvalho P, Gupta ML, Jr., Hoyt MA, Pellman D. Cell cycle control of kinesin-mediated transport of Bik1 (CLIP-170) regulates microtubule stability and dynein activation. Dev Cell. 2004;6:815–829. doi: 10.1016/j.devcel.2004.05.001. [DOI] [PubMed] [Google Scholar]
- Cassimeris L, Gard D, Tran PT, Erickson HP. XMAP215 is a long thin molecule that does not increase microtubule stiffness. J Cell Sci. 2001;114:3025–3033. doi: 10.1242/jcs.114.16.3025. [DOI] [PubMed] [Google Scholar]
- Choi J, Chen J, Schreiber SL, Clardy J. Structure of the FKBP12-rapamycin complex interacting with the binding domain of human FRAP. Science. 1996;273:239–242. doi: 10.1126/science.273.5272.239. [DOI] [PubMed] [Google Scholar]
- Choi JH, Bertram PG, Drenan R, Carvalho J, Zhou HH, Zheng XF. The FKBP12-rapamycin-associated protein (FRAP) is a CLIP-170 kinase. EMBO Rep. 2002;3:988–994. doi: 10.1093/embo-reports/kvf197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cingolani G, Petosa C, Weis K, Muller CW. Structure of importin-beta bound to the IBB domain of importin-alpha. Nature. 1999;399:221–229. doi: 10.1038/20367. [DOI] [PubMed] [Google Scholar]
- Cullen CF, Deak P, Glover DM, Ohkura H. mini spindles: A gene encoding a conserved microtubule-associated protein required for the integrity of the mitotic spindle in Drosophila. J Cell Biol. 1999;146:1005–1018. doi: 10.1083/jcb.146.5.1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai A, Mitchison TJ. Microtubule polymerization dynamics. Annu Rev Cell Dev Biol. 1997;13:83–117. doi: 10.1146/annurev.cellbio.13.1.83. [DOI] [PubMed] [Google Scholar]
- Diamantopoulos GS, Perez F, Goodson HV, Batelier G, Melki R, Kreis TE, Rickard JE. Dynamic localization of CLIP-170 to microtubule plus ends is coupled to microtubule assembly. J Cell Biol. 1999;144:99–112. doi: 10.1083/jcb.144.1.99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dougherty GW, Adler HJ, Rzadzinska A, Gimona M, Tomita Y, Lattig MC, Merritt RC, Jr., Kachar B. CLAMP, a novel microtubule-associated protein with EB-type calponin homology. Cell Motil Cytoskeleton. 2005;62:141–156. doi: 10.1002/cm.20093. [DOI] [PubMed] [Google Scholar]
- Folker ES, Baker BM, Goodson HV. Interactions between CLIP-170, tubulin, and microtubules: implications for the mechanism of Clip-170 plus-end tracking behavior. Mol Biol Cell. 2005;16:5373–5384. doi: 10.1091/mbc.E04-12-1106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graf R, Daunderer C, Schliwa M. Dictyostelium DdCP224 is a microtubule-associated protein and a permanent centrosomal resident involved in centrosome duplication. J Cell Sci. 2000;113(Pt 10):1747–1758. doi: 10.1242/jcs.113.10.1747. [DOI] [PubMed] [Google Scholar]
- Hayashi I, Ikura M. Crystal structure of the amino-terminal microtubule-binding domain of end-binding protein 1 (EB1) J Biol Chem. 2003;278:36430–36434. doi: 10.1074/jbc.M305773200. [DOI] [PubMed] [Google Scholar]
- Hayashi I, Wilde A, Mal TK, Ikura M. Structural basis for the activation of microtubule assembly by the EB1 and p150Glued complex. Mol Cell. 2005;19:449–460. doi: 10.1016/j.molcel.2005.06.034. [DOI] [PubMed] [Google Scholar]
- Honnappa S, John CM, Kostrewa D, Winkler FK, Steinmetz MO. Structural insights into the EB1-APC interaction. Embo J. 2005;24:261–269. doi: 10.1038/sj.emboj.7600529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honnappa S, Okhrimenko O, Jaussi R, Jawhari H, Jelesarov I, Winkler FK, Steinmetz MO. Key interaction modes of dynamic +TIP networks. Mol Cell. 2006;23:663–671. doi: 10.1016/j.molcel.2006.07.013. [DOI] [PubMed] [Google Scholar]
- Hoogenraad CC, Akhmanova A, Grosveld F, De Zeeuw CI, Galjart N. Functional analysis of CLIP-115 and its binding to microtubules. J Cell Sci. 2000;113(Pt 12):2285–2297. doi: 10.1242/jcs.113.12.2285. [DOI] [PubMed] [Google Scholar]
- Horio T, Hotani H. Visualization of the dynamic instability of individual microtubules by dark-field microscopy. Nature. 1986;321:605–607. doi: 10.1038/321605a0. [DOI] [PubMed] [Google Scholar]
- Kerssemakers JW, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340–1343. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- Lansbergen G, Akhmanova A. Microtubule plus end: a hub of cellular activities. Traffic. 2006;7:499–507. doi: 10.1111/j.1600-0854.2006.00400.x. [DOI] [PubMed] [Google Scholar]
- Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- Li S, Finley J, Liu ZJ, Qiu SH, Chen H, Luan CH, Carson M, Tsao J, Johnson D, Lin G, et al. Crystal structure of the cytoskeleton-associated protein glycine-rich (CAP-Gly) domain. J Biol Chem. 2002;277:48596–48601. doi: 10.1074/jbc.M208512200. [DOI] [PubMed] [Google Scholar]
- Ligon LA, Shelly SS, Tokito MK, Holzbaur EL. Microtubule binding proteins CLIP-170, EB1, and p150Glued form distinct plus-end complexes. FEBS Lett. 2006;580:1327–1332. doi: 10.1016/j.febslet.2006.01.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H, Khodjakov A, Rieder CL. Drosophila CLASP is required for the incorporation of microtubule subunits into fluxing kinetochore fibres. Nat Cell Biol. 2005;7:42–47. doi: 10.1038/ncb1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- Mimori-Kiyosue Y, Shiina N, Tsukita S. The dynamic behavior of the APC-binding protein EB1 on the distal ends of microtubules. Curr Biol. 2000;10:865–868. doi: 10.1016/s0960-9822(00)00600-x. [DOI] [PubMed] [Google Scholar]
- Mino T, Yuasa U, Nakamura F, Naka M, Tanaka T. Two distinct actin-binding sites of smooth muscle calponin. Eur J Biochem. 1998;251:262–268. doi: 10.1046/j.1432-1327.1998.2510262.x. [DOI] [PubMed] [Google Scholar]
- Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- Moores CA, Milligan RA. Lucky 13-microtubule depolymerisation by kinesin-13 motors. J Cell Sci. 2006;119:3905–3913. doi: 10.1242/jcs.03224. [DOI] [PubMed] [Google Scholar]
- Nakaseko Y, Goshima G, Morishita J, Yanagida M. M phase-specific kinetochore proteins in fission yeast: microtubule-associating Dis1 and Mtc1 display rapid separation and segregation during anaphase. Curr Biol. 2001;11:537–549. doi: 10.1016/s0960-9822(01)00155-5. [DOI] [PubMed] [Google Scholar]
- Niethammer P, Kronja I, Kandels-Lewis S, Rybina S, Bastiaens P, Karsenti E. Discrete states of a protein interaction network govern interphase and mitotic microtubule dynamics. PLoS Biol. 2007;5:e29. doi: 10.1371/journal.pbio.0050029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez F, Diamantopoulos GS, Stalder R, Kreis TE. CLIP-170 highlights growing microtubule ends in vivo. Cell. 1999;96:517–527. doi: 10.1016/s0092-8674(00)80656-x. [DOI] [PubMed] [Google Scholar]
- Peris L, Thery M, Faure J, Saoudi Y, Lafanechere L, Chilton JK, Gordon-Weeks P, Galjart N, Bornens M, Wordeman L, et al. Tubulin tyrosination is a major factor affecting the recruitment of CAP-Gly proteins at microtubule plus ends. J Cell Biol. 2006;174:839–849. doi: 10.1083/jcb.200512058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierre P, Pepperkok R, Kreis TE. Molecular characterization of two functional domains of CLIP-170 in vivo. J Cell Sci. 1994;107(Pt 7):1909–1920. doi: 10.1242/jcs.107.7.1909. [DOI] [PubMed] [Google Scholar]
- Pierre P, Scheel J, Rickard JE, Kreis TE. CLIP-170 links endocytic vesicles to microtubules. Cell. 1992;70:887–900. doi: 10.1016/0092-8674(92)90240-d. [DOI] [PubMed] [Google Scholar]
- Popov AV, Pozniakovsky A, Arnal I, Antony C, Ashford AJ, Kinoshita K, Tournebize R, Hyman AA, Karsenti E. XMAP215 regulates microtubule dynamics through two distinct domains. Embo J. 2001;20:397–410. doi: 10.1093/emboj/20.3.397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinlan ME, Heuser JE, Kerkhoff E, Mullins RD. Drosophila Spire is an actin nucleation factor. Nature. 2005;433:382–388. doi: 10.1038/nature03241. [DOI] [PubMed] [Google Scholar]
- Rogers SL, Rogers GC, Sharp DJ, Vale RD. Drosophila EB1 is important for proper assembly, dynamics, and positioning of the mitotic spindle. J Cell Biol. 2002;158:873–884. doi: 10.1083/jcb.200202032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samsonov A, Yu JZ, Rasenick M, Popov SV. Tau interaction with microtubules in vivo. J Cell Sci. 2004;117:6129–6141. doi: 10.1242/jcs.01531. [DOI] [PubMed] [Google Scholar]
- Sandblad L, Busch KE, Tittmann P, Gross H, Brunner D, Hoenger A. The Schizosaccharomyces pombe EB1 homolog Mal3p binds and stabilizes the microtubule lattice seam. Cell. 2006;127:1415–1424. doi: 10.1016/j.cell.2006.11.025. [DOI] [PubMed] [Google Scholar]
- Sept D, Baker NA, McCammon JA. The physical basis of microtubule structure and stability. Protein Sci. 2003;12:2257–2261. doi: 10.1110/ps.03187503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slep KC, Rogers SL, Elliott SL, Ohkura H, Kolodziej PA, Vale RD. Structural determinants for EB1-mediated recruitment of APC and spectraplakins to the microtubule plus end. J Cell Biol. 2005;168:587–598. doi: 10.1083/jcb.200410114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spittle C, Charrasse S, Larroque C, Cassimeris L. The interaction of TOGp with microtubules and tubulin. J Biol Chem. 2000;275:20748–20753. doi: 10.1074/jbc.M002597200. [DOI] [PubMed] [Google Scholar]
- Srayko M, Kaya A, Stamford J, Hyman AA. Identification and characterization of factors required for microtubule growth and nucleation in the early C. elegans embryo. Dev Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- Sutherland-Smith AJ, Moores CA, Norwood FL, Hatch V, Craig R, Kendrick-Jones J, Lehman W. An atomic model for actin binding by the CH domains and spectrin-repeat modules of utrophin and dystrophin. J Mol Biol. 2003;329:15–33. doi: 10.1016/s0022-2836(03)00422-4. [DOI] [PubMed] [Google Scholar]
- Tirnauer JS, Salmon ED, Mitchison TJ. Microtubule plus-end dynamics in Xenopus egg extract spindles. Mol Biol Cell. 2004;15:1776–1784. doi: 10.1091/mbc.E03-11-0824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tournebize R, Popov A, Kinoshita K, Ashford AJ, Rybina S, Pozniakovsky A, Mayer TU, Walczak CE, Karsenti E, Hyman AA. Control of microtubule dynamics by the antagonistic activities of XMAP215 and XKCM1 in Xenopus egg extracts. Nat Cell Biol. 2000;2:13–19. doi: 10.1038/71330. [DOI] [PubMed] [Google Scholar]
- van Breugel M, Drechsel D, Hyman A. Stu2p, the budding yeast member of the conserved Dis1/XMAP215 family of microtubule-associated proteins is a plus end-binding microtubule destabilizer. J Cell Biol. 2003;161:359–369. doi: 10.1083/jcb.200211097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanBuren V, Odde DJ, Cassimeris L. Estimates of lateral and longitudinal bond energies within the microtubule lattice. Proc Natl Acad Sci U S A. 2002;99:6035–6040. doi: 10.1073/pnas.092504999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez RJ, Gard DL, Cassimeris L. XMAP from Xenopus eggs promotes rapid plus end assembly of microtubules and rapid microtubule polymer turnover. J Cell Biol. 1994;127:985–993. doi: 10.1083/jcb.127.4.985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan KT. TIP maker and TIP marker; EB1 as a master controller of microtubule plus ends. J Cell Biol. 2005;171:197–200. doi: 10.1083/jcb.200509150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaughan PS, Miura P, Henderson M, Byrne B, Vaughan KT. A role for regulated binding of p150(Glued) to microtubule plus ends in organelle transport. J Cell Biol. 2002;158:305–319. doi: 10.1083/jcb.200201029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei RR, Al-Bassam J, Harrison SC. The Ndc80/HEC1 complex is a contact point for kinetochore-microtubule attachment. Nat Struct Mol Biol. 2007;14:54–59. doi: 10.1038/nsmb1186. [DOI] [PubMed] [Google Scholar]
- Wittmann T, Waterman-Storer CM. Spatial regulation of CLASP affinity for microtubules by Rac1 and GSK3beta in migrating epithelial cells. J Cell Biol. 2005;169:929–939. doi: 10.1083/jcb.200412114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolyniak MJ, Blake-Hodek K, Kosco K, Hwang E, You L, Huffaker TC. The regulation of microtubule dynamics in Saccharomyces cerevisiae by three interacting plus-end tracking proteins. Mol Biol Cell. 2006;17:2789–2798. doi: 10.1091/mbc.E05-09-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.