Abstract
The association of abnormal spermatogenesis in men with Y chromosome deletions suggests that genes important for spermatogenesis have been removed from these individuals. Recently, genes encoding two putative RNA-binding proteins (RBM and DAZ/SPGY) have been mapped to two different regions of the human Y chromosome. Both of these genes encode proteins that contain a single RNA recognition motif and a (different) internally repeating sequence. Y-linked RBM homologues are found in all mammalian species. We have raised an antiserum to RBM and used it to show that RBM is a nuclear protein expressed in fetal, prepubertal, and adult male germ cells. The distribution of RBM protein in the adult correlates with the pattern of transcriptional activity in spermatogenesis, suggesting that RBM is involved in the nuclear metabolism of newly synthesized RNA. RBM sequences are found on both arms of the Y chromosome making genotype–phenotype correlations difficult for this gene family. To address the location of the functional genes and the consequences of their deletion, we examined a panel of men with Y chromosome deletions and known testicular pathologies using this antiserum. This approach enabled us to map a region of the Y chromosome essential for RBM expression. In the absence of detectable RBM expression we see stages of germ cell development up to early meiosis, but not past this point into the haploid phase of spermatogenesis.
The Y chromosome is the smallest human chromosome, contributing only 2–3% of the haploid genome, and is restricted to males. At the cytogenetic level, the Y chromosome has a long and short arm, demarcated by a centromeric region essential for chromosome segregation. Only small regions homologous to the X chromosome at the tips of the Y chromosome undergo genetic recombination—the pseudoautosomal regions. The remainder of the chromosome is the only haploid compartment of the human genome. This has evolutionary consequences and also facilitates deletion mapping, since deletions are viable and any mutations cannot usually be masked by a normal allele.
The presence of individuals with deleted Y chromosomes has been useful for genetic mapping. Studies of these individuals has allowed deletion intervals to be ordered along the chromosome and the relative position of markers to be mapped by molecular techniques [Southern blotting or sequence-tagged sites (STSs)] to be localized, in addition to the assembly of an ordered yeast artificial chromosome contig (1–3). These genetic maps have subsequently been used to localize genes encoded by the Y chromosome, the absence of which are associated with various pathologies (4–6). Visible deletions of the long arm were associated with azoospermia. This observation led to the prediction of a gene or genes encoded within this interval called the azoospermia factor (AZF; ref. 7–12). Further work suggests the presence of multiple spermatogenesis loci on the Y chromosome, which map to regions that have been termed AZFa, AZFb, and AZFc (13). AZFa has previously been called the JOLAR region (9) and AZFc the “minimal AZF region” (12). Microdeletions of these loci are associated with defects in spermatogenesis leading to either greatly reduced numbers of sperm (oligozoospermia) or to their complete absence (azoospermia).
Recently, genes encoding two RNA-binding proteins, called RBM and DAZ/SPGY, have been mapped to the Y chromosome (12, 14, 15) and they are candidate AZF genes. Both these proteins contain a single RNA recognition motif, which is a protein motif found in a diverse array of RNA-binding proteins (16) and has been shown in some examples to interact with RNA. Both RBM and DAZ/SPGY proteins also contain a (different) internally repeating structure. DAZ/SPGY is present in, at most, a few copies on the Y chromosome, which are frequently deleted in men with spermatogenic defects, and maps to AZFc (12, 13). The multigene nature of RBM in humans (14) and in mice (17, 18) has complicated its functional analysis. Deletions of some Rbm copies in the Yd mice occur, but these mice are XY females, so effects on spermatogenesis could not be determined (18). To investigate the function of RBM, we have developed an antiserum that specifically recognizes the protein and used it to examine its pattern of expression in both the adult and fetal testis. We have also used it to examine RBM expression in the testis of men with defined Y chromosome deletions. The use of antibody probes to identify the chromosomal locations of active members of multigene families is a novel approach that could be usefully applied to other situations.
MATERIALS AND METHODS
Generation of Antibodies Against RBM.
The SRGY box of pMK5 was PCR amplified using oligonucleotides 5′-AAA AAA ACT CGA GAA CGA TGG AAA TCA TCC AAG T and 5′-AAA AAA AGG TAC CAT GAT TTC TAT ATC CTC TAG A. The PCR product was gel-purified, digested with XhoI and KpnI (restriction sites underlined), and ligated into pRSETA. The resulting plasmid, pH2, was transformed into Escherichia coli BLR cells (Invitrogen), and a polypeptide of ≈25 kDa was induced by the addition of isopropyl β-d-thiogalactoside. This polypeptide was purified as an inclusion body after cell lysis in 20 mM sodium phosphate/500 mM sodium chloride, pH 7.8, solubilized in 50 mM Tris/2 mM EDTA, pH 8.0, and then injected into a rabbit as an emulsion with TitreMax adjuvant (CytRx, Norcross, GA). The rabbit was boosted after 5, 10, and 20 weeks and test-bled 12–14 days after each injection, with a final bleeding 23 weeks after the initial injection. Antibodies in the blood taken after the second boost were affinity-purified against the immunizing protein on an Affigel 10 column (Bio-Rad) according to the manufacturers instructions, and this purified antiserum was used for further experiments.
Immunohistochemistry.
Surgically removed testes were fixed in Bouins’ solution and embedded in paraffin wax. Sections were processed for immunohistochemistry according to standard procedures, and antigens were revealed by microwave treatment in 0.01 M citrate buffer (pH 6.0; refs. 19 and 20). A 1:500 dilution of affinity-purified anti-RBM antibody and a 1:400 dilution of biotin-conjugated swine anti-rabbit antibody (Dako) were used for detection, followed by an avidin–biotin–horseradish peroxidase (ABC–HRP) system (Dako) and diaminobenzidine (DAB; Sigma). Sections were briefly counterstained in Harris hematoxylin and mounted in DPX mounting medium (BDH; Merck).
Mapping of Patient Deletion Breakpoints.
STSs defining microdeletion breakpoints were mapped by Qureshi et al. (21) The breakpoints of patients BITRA and HORIS were mapped by assaying the presence of various STSs, as described by Volrath et al. (2).
RESULTS
Subcellular Localization of RBM.
RBM is homologous to the autosomal gene that encodes heterogeneous ribonucleoprotein (hnRNP) G [which is a member of the large family of hnRNPs associated with nuclear polyadenylylated RNA (22, 23)], showing 76% similarity and 60% identity at the amino acid level. While regions of homology are spread fairly evenly throughout the whole molecule, RBM contains four tandem repeats of a 37-aa peptide called the SRGY box not present in hnRNP G. To generate antibodies that are specific for RBM, we expressed this SRGY box in E. coli and used the polypeptide to immunize rabbits. The resulting antiserum detected a major testis-specific protein by Western blotting, which corresponded in size with the predicted molecular weight of RBM (data not shown).
To define which cell types express RBM and its subcellular location, we used the anti-RBM antiserum to probe histological sections of adult human testes (Fig. 1a). Only germ cells were stained with the antisera; somatic cells in the seminiferous tubules (Sertoli and myoid cells) were not stained, and neither were interstitial Leydig cells. Strong nuclear staining was observed in both type A and B spermatogonia, spermatocytes, and round spermatids. In contrast, both germ cells and somatic cells were labeled with a monoclonal antibody that recognizes Sm, an antigen present on a number of nuclear RNPs (refs. 24 and 25; Fig. 1b). Importantly, the expression pattern of RBM correlates with the pattern of relative transcriptional activity in germ cells during spermatogenesis. No RBM protein is detectable in elongating spermatids that are transcriptionally quiescent (26, 27). This is consistent with a role for RBM in the nuclear metabolism of newly synthesized RNA—e.g., in pre-mRNA splicing, polyadenylylation, or nonspecific binding with nascent RNA.
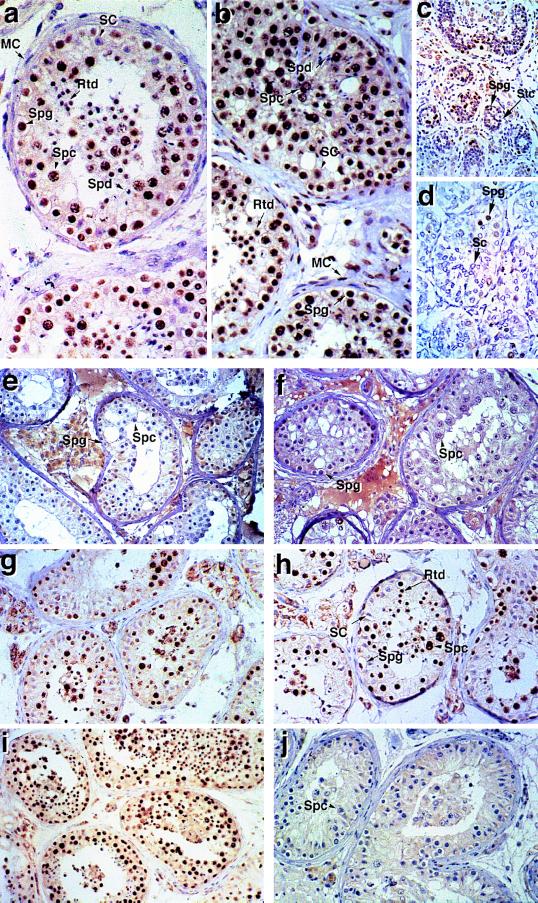
Figure 1.
Immunocytochemical localization of RBM in testes from normal fertile men and men with Y chromosome deletions. (a) Normal adult testis probed with anti-RBM antiserum. Examples of each cell type are identified. Spermatogonia (Spg, at tubule periphery), spermatocytes (Spc, note the condensed chromosomes visible during meiosis), and round spermatids (Rtd, smaller nuclei with a haploid DNA content) are labeled by the anti-RBM antiserum, while elongating spermatids (Spd), Sertoli cells (SC), and myoid cells (MC) are unlabeled. (b) Normal adult testis probed with the anti-Sm monoclonal antibody Y12, showing all nuclei apart from elongating spermatids are stained. Examples of each cell type are identified as in a. (c) Fetal testis (24 weeks after conception) probed with the anti-RBM antiserum. Primordial spermatogonia (Spg) or gonocytes are stained by the anti-RBM antibody, while sustenticular cells (Stc) are not. (d) Prepubertal (2 days after birth) testis probed with the anti-RBM antiserum, showing spermatogonia are stained while Sertoli cells (Sc) are not. (e) Testis section from patient H139 probed with the anti-RBM antiserum showing that spermatogonia and spermatocytes do not express RBM in this individual. (f) Testis section from patient GLIPS probed with the anti-RBM antiserum, showing that spermatogonia and spermatocytes do not express RBM in this individual. (g) Testis section from patient NICKEI probed with the anti-RBM antiserum, showing positive RBM staining of germ cell nuclei. (h) Testis section from patient HAMIL probed with the anti-RBM antiserum, showing positive RBM staining of germ cell nuclei, while Sertoli cells are not labeled by the antibody. (i) Testis section from patient SAYER probed with the anti-RBM antiserum, showing positive RBM staining of germ cell nuclei. (j) Testis section from patient BITRA probed with the anti-RBM antiserum, showing that spermatogonia and spermatocytes do not express RBM in this individual. In all cases, antibody binding was visualized using DAB as a substrate to generate a brown color. Sections were counterstained with Harris hematoxylin, which stains nuclei blue. No germ cell nuclear staining was observed when sections were stained with preimmune serum from the same rabbit used to raise the antiserum, and staining was quantitatively reduced to near zero by preincubation with the immunizing polypeptide (data not shown). In all cases, weak interstitial labeling was observed, which therefore must represent a nonspecific interaction. This is particularly clear in e and f, which were developed longer to show up any RBM staining.
As a number of transcripts encoding proteins essential for spermatogenesis are alternatively processed in the testis, such as CREM (28–30), RNA processing defects might therefore lead indirectly to spermatogenic defects. The RNAs to which RBM putatively binds may either not be restricted to specific stages of spermatogenesis, or RBM may have different target RNAs in different germ cell types. In the latter case, the specificity of RBM may be modulated by other stage-specific factors. Consistent with these possibilities, RBM was also strongly expressed in the nuclei of fetal germ cells at 16, 18, and 24 weeks gestation (data not shown and Fig. 1c) and in the testis of a 2-day-old boy who died neonatally (Fig. 1d). Whereas the bulk of cells within the seminiferous tubules in the adult testis are germ cells, in fetal and early neonatal life, these are arrested at G1 of mitosis and occupy only a small proportion of the seminiferous cords. This is reflected in the reduced number of positively staining cells with the anti-RBM antiserum at these ages.
A Subregion of Y Chromosome Interval 6 Is Critical for RBM Expression.
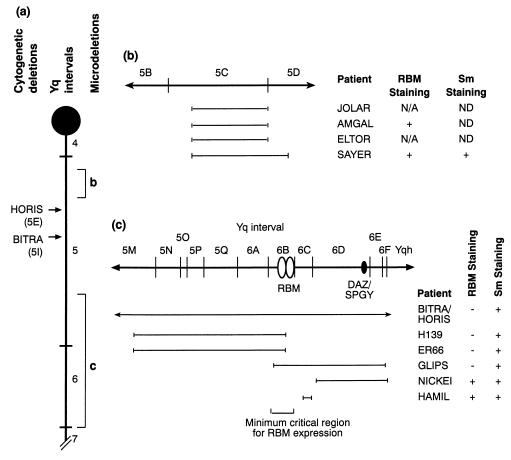
RBM is a multigene family, with copies (some of which are transcribed pseudogenes) mapping to different regions of the Y chromosome, on both arms (12, 14). Large deletions can occur, leading to a number of genes being removed from the genome simultaneously, among them copies of the RBM gene, and their precise breakpoints can be mapped on the basis of their content of STSs and other markers (1, 2). These deletion intervals have been numbered and ordered on the Y chromosome (refs. 2 and 12; Fig. 2). In addition, microdeletions of three regions have been described in men with impaired spermatogenesis, called the AZFa region (subinterval 5C), the AZFb region (including distal subinterval 5 and proximal subinterval 6), and the AZFc region (between subinterval 6D and the heterochromatic region of the Y, Yqh) (refs. 9 and 12–14; Fig. 2).
Figure 2.
Map of the Y chromosome long arm. (a) Map of the Y chromosome long arm (Yq) showing the position of deletion intervals 4–7 (12) and the centromere (black circle). The positions of the BITRA and HORIS deletion breakpoints are shown as arrows (with map locations in parentheses), which result in the deletion of the entire Y chromosome distal to this point (i.e., the rest of interval 5 and intervals 6 and 7). Regions of the Y chromosome long arm frequently microdeleted in infertile men are labeled b and c, and these are shown in more detail in b and c, respectively. (b) More detailed map of the subintervals 5B–5D of the Y chromosome, which contain the AZFa locus (13), showing underneath the extent of microdeletions in patients missing this region of the Y chromosome alongside the name of each patient, and a summary of both RBM and Sm expression in the germ cells of these men. The size of the microdeletion shown corresponds to the number of missing STSs (see Table 1 and ref. 12). The rest of the Y chromosome outside the region indicated to be deleted is intact in these men. Neither JOLAR or ELTOR have germ cells, and so RBM expression cannot be visualized (labeled N/A). Testes sections not probed for Sm are labeled ND. (c) Detail of distal interval 5/interval 6 showing the the locations of the mapped RBM and DAZ/SPGY genes and the position of microdeletions in this region, along with the names of the corresponding patients. The NICKEI microdeletion defines the AZFc locus, while the H139 and ER66 microdeletions define the AZFb locus (13). The expression of RBM and Sm in germ cells in each of these patients is summarized alongside the name of the patient, and the minimum critical region for RBM expression is indicated underneath. BITRA and HORIS are cytological deletions which remove this entire region (see a).
To determine if any of these regions of the Y chromosome are required for quantitative RBM expression, we screened for RBM expression in testis sections from a panel of patients with microdeletions in these regions (Table 1). Both patients ER66 and H139, men with interstitial deletions that correspond to AZFb and who have intact DAZ/SPGY genes (Fig. 2), have RBM-negative germ cells (Fig. 1e and data not shown). Additionally, RBM expression in germ cells in patient GLIPS, who has a slightly overlapping microdeletion (between subinterval 6A and the heterochromatic region, including DAZ/SPGY), was barely detectable or zero (Fig. 1f and data not shown). These results suggest that the AZFb region is critical for RBM expression. However, patients HAMIL and NICKEI (Fig. 1 g and h), who have microdeletions in the AZFc region (in both cases removing DNA between subinterval 6C and the heterochromatic region Yqh) and are deleted for the DAZ/SPGY gene, express RBM in their germ cells. Hence, the AZFc region is not important for quantitative RBM expression. Both patients SAYER (Fig. 1i) and AMGAL (data not shown), who have microdeletions in the AZFa region and show some spermatogenesis, stained positively for RBM. This implies that the region required for RBM expression is outside the AZFa region. Consistent with these data, patients HORIS and BITRA (31, 32), men with cytologically detectable terminal deletions of Yq from interval 5I (which includes both AZFb and AZFc regions; Fig. 2) were also RBM-negative (Fig. 1j). No RBM staining was observed in patients JOLAR and ELTOR (data not shown), since these men lacked germ cells. In control experiments, germ cells in testis sections from men not expressing RBM stained positively for the Sm antigen, showing that their nuclear antigens were accessible to antibody binding (data not shown; summarized in Fig. 2).
Table 1.
Phenotype and genotype of men used in this study.
| Patient | Mean sperm count (×106/ml) | Johnsen score | Testicular histology | Marker defining breakpoint | Ref(s). |
|---|---|---|---|---|---|
| BITRA | Azoospermic | 2.6 | Most tubules contain only Sertoli cells Minority show limited spermatogonial proliferation up to spermatocyte stage, atrophy | sY101 (Del Yq) | 31, 32, and M. Kent-First, personal communication |
| HORIS | Azoospermic | 2.2 | Similar to BITRA | sY89 (Del Yq) | This work |
| JOLAR | Azoospermic | 2 | SCO | sY84–sY86 | 21 |
| ELTOR | Azoospermic | 2 | SCO | sY84–sY86 | 21 |
| SAYER | 4.4 | 7.7 | Few tubules contain either only Sertoli cells or spermatogonia, spermatocytes, or spermatids; most contain small to moderate numbers of spermatogonia | sY84–sY87 | 21 |
| AMGAL | Azoospermic | 2 | Similar to BITRA | sY84–ND | 21 |
| NICKEI | <0.01 | 6.2 | Severely diminished spermatogenesis with partial arrest at the primary spermatocyte and spermatid stage | sY232–sY158 | 21 |
| HAMIL | 0.6 | 5.1 | Most tubules SCO. Minority show spermatogenesis to the spermatozoan stage. | sY152 | 21 |
| GLIPS | Azoospermic | 2 | Similar to BITRA | sY153–sY158 | This work |
| H139 | Azoospermic | 4.9 | Pachytene arrest | sY117–Y367/A | 13 |
| ER66 | Azoospermic | 4.6 | Pachytene arrest | sY117–Y367/A | This work |
STSs were assayed as described by Volrath et al. (2). The pair of markers defining the breakpoint (negative in these patients) are given except for BITRA and HORIS, who have deleted Y chromosomes, which remove the chromosome distal to the breakpoint including the distal heterochromatin, Yqh. Johnsen scores (33) were assessed independently from hematoxylin- and eosin-stained slides. Johnsen scores are a numerical score from 1 to 10 that assess the quality of spermatogenesis, with 10 being full spermatogenesis. ND, not determined; SCO, Sertoli cell only.
DISCUSSION
Taken as a whole, these results indicate that a region of the Y chromosome long arm in distal interval 5/proximal interval 6 is essential for quantitative RBM expression, and this is coincident with the AZFb locus, which has been implicated as important for normal spermatogenesis (13). The most straightforward interpretation of these results is that the most active copy(ies) of the RBM gene are located within this critical region. Consistent with this hypothesis, RBM genes have been mapped within proximal deletion interval 6 (refs. 12 and 34; Fig. 2). An implication of this result is that RBM genes on the short arm and proximal long arm of the Y chromosome do not make quantitative contributions to the steady-state level of RBM protein. Men with AZFb deletions do contain other copies of RBM, so despite having such a dramatic effect on RBM expression, disruption of RBM would not have been easily detected by Southern blot or PCR assays. These data imply that the RBM gene family does not contain functionally redundant members with disparate Y chromosome locations, but rather the active gene(s) is located in a comparatively small region of the chromosome. In addition to physical deletion, position effects (35) in microdeletion patients like GLIPS [and indeed KUPAU and KLARD, who have not been biopsied (14)] might functionally inactivate this region by placing it adjacent to the heterochromatin present at the end of the Y chromosome long arm (Yqh). Our data further indicate that deletion of either the AZFa or AZFc region (13) has no detectable effect on RBM expression.
These results help to define the role of the RBM protein in human germ cell development. The expression of RBM in the majority of germ cells in the adult testis, as well as in the fetal and prepubertal testis, is consistent with a role for RBM throughout germ cell development. However, since germ cell development up to meiosis was observed in the absence of quantitative RBM expression, RBM is not absolutely essential for the development up to the meiotic stages of spermatogenesis, although it might be required for efficient development up to this stage. However, RBM may be required either for the completion of meiosis or subsequent haploid stages, although we cannot exclude the possibility that the effect of RBM deletion is earlier and the arrest we see is a more generalized later consequence. The conservation of Y-linked RBM genes in all mammals implies an important function in fertility (14).
Many patients reported in this study are also deleted for DAZ/SPGY. Disruption of the Drosophila autosomal homologue of DAZ/SPGY results in meiotic arrest (36). Although men deleted for DAZ/SPGY alone can progress past the meiotic stage of spermatogenesis (12), deletion of DAZ/SPGY may be contributing to their spermatogenic impairment. In contrast, both men who are DAZ/SPGY gene-positive but who do not express RBM protein are arrested in meiosis, as is GLIPS, who is deleted for both DAZ/SPGY and RBM. This suggests a distinct role for the two proteins with a more severe phenotype resulting from RBM deletion. However, the number of patients is small, and so the phenotypic data should be interpreted with caution.
Studies on hnRNP expression in mouse testes have shown that hnRNP A1 is only expressed in spermatogonia, while no hnRNP expression was detected in round spermatids (37), although these cells continue to be active in RNA synthesis. Hence, the role of RBM (and/or DAZ/SPGY), which is expressed throughout spermatogenesis, might be to functionally replace these molecules in germ cell nuclei.
Acknowledgments
Dr. J. Keeling is thanked for supplying the fetal and prepubertal testis used in this study, and Professors J. Steitz and A. I. Lamond are thanked for supplying monoclonal antibody Y12. Dr. Shona Kerr, Dr. David Kipling, and Prof. Nick Hastie are thanked for constructive comments on the manuscript. This work was supported by the Medical Research Council.
ABBREVIATIONS
- STS
sequence-tagged site
- AZF
azoospermia factor
- hnRNP
heterogeneous nuclear ribonucleoprotein
References
- 1.Vergnaud G, Page D C, Simmler M-C, Brown L, Rouyer F, Noel B, Botstein D, de la Chapelle A, Weissenbach J. Am J Hum Genet. 1986;38:109–124. [PMC free article] [PubMed] [Google Scholar]
- 2.Volrath D, Foote S, Hilton A, Brown L G, Beer-Romero P, Bogan J S, Page D C. Science. 1992;258:52–59. doi: 10.1126/science.1439769. [DOI] [PubMed] [Google Scholar]
- 3.Foote S, Volrath D, Hilton A, Page D C. Science. 1992;258:60–66. doi: 10.1126/science.1359640. [DOI] [PubMed] [Google Scholar]
- 4.Fisher E M C, Beer-Romero P, Brown L G, Ridley A, Mcneil J A, Lawrence J B, Willard H F, Bieber F R, Page D C. Cell. 1990;63:1205–1218. doi: 10.1016/0092-8674(90)90416-c. [DOI] [PubMed] [Google Scholar]
- 5.Sinclair A H, Berta P, Palmer M S, Hawkins J R, Griffiths B L, Smith M J, Foster J W, Frischauf A-M, Lovell-Badge R, Goodfellow P N. Nature (London) 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- 6.Salo P, Kaariainen H, Page D C, de la Chapelle A. Hum Genet. 1995;95:283–286. doi: 10.1007/BF00225194. [DOI] [PubMed] [Google Scholar]
- 7.Tiepolo L, Zuffardi O. Hum Genet. 1976;34:119–124. doi: 10.1007/BF00278879. [DOI] [PubMed] [Google Scholar]
- 8.Andersson M, Page D, Pettay D, Subrt I, Turleau C, de Grouchy J, de la Chapelle A. Hum Genet. 1988;79:2–7. doi: 10.1007/BF00291700. [DOI] [PubMed] [Google Scholar]
- 9.Ma K, Sharkey A, Kirsch S, Vogt P, Keil R, Hargreave T B, McBeath S, Chandley A C. Hum Mol Genet. 1992;1:29–33. doi: 10.1093/hmg/1.1.29. [DOI] [PubMed] [Google Scholar]
- 10.Vogt P, Chandley A C, Hargreave T B, Keil R, Ma K, Sharkey A. Hum Genet. 1992;89:491–496. doi: 10.1007/BF00219172. [DOI] [PubMed] [Google Scholar]
- 11.Chandley A C, Cooke H J. Hum Mol Genet. 1994;3:1449–1452. doi: 10.1093/hmg/3.suppl_1.1449. [DOI] [PubMed] [Google Scholar]
- 12.Reijo R, Lee T Y, Salo P, Alagappan R, Brown L G, Rosenberg M, Rozen S, Jaffe T, Straus D, Hovatta O, de la Chapelle A, Silber S, Page D C. Nat Genet. 1995;10:383–393. doi: 10.1038/ng0895-383. [DOI] [PubMed] [Google Scholar]
- 13.Vogt P H, Edelmann A, Kirsch S, Henegariu O, Hirschmann P, et al. Hum Mol Genet. 1996;5:933–943. doi: 10.1093/hmg/5.7.933. [DOI] [PubMed] [Google Scholar]
- 14.Ma K, Inglis J D, Sharkey A, Bickmore W A, Hill R E, Prosser E J, Speed R M, Thomson E J, Jobling M, Taylor K, Wolfe J, Cooke H J, Hargreave T B, Chandley A C. Cell. 1993;75:1287–1295. doi: 10.1016/0092-8674(93)90616-x. [DOI] [PubMed] [Google Scholar]
- 15.Maiwald R, Seebacher T, Edelmann A, Hirschmann P, Kohler M R, Kirsch S, Vogt P. Cytogenet Cell Genet. 1996;73:33–76. [Google Scholar]
- 16.Kenan D J, Query C C, Keene J D. Trends Biochem Sci. 1991;16:214–220. doi: 10.1016/0968-0004(91)90088-d. [DOI] [PubMed] [Google Scholar]
- 17.Elliott D J, Ma K, Kerr S M, Thakrar R, Speed R, Chandley A C, Cooke H. Hum Mol Genet. 1996;5:869–874. doi: 10.1093/hmg/5.7.869. [DOI] [PubMed] [Google Scholar]
- 18.Laval S H, Glenister P H, Rasberry C, Thornton C E, Mahadevaiah S K, Cooke H J, Burgoyne P S, Cattanach B M. Proc Natl Acad Sci USA. 1995;92:10403–10407. doi: 10.1073/pnas.92.22.10403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bremner W J, Millar M R, Sharpe R M, Saunders P T K. Endocrinology. 1994;135:1227–1234. doi: 10.1210/endo.135.3.8070367. [DOI] [PubMed] [Google Scholar]
- 20.Shi S, Chaiwun B, Young L, Cote R J, Taylor C R. J Histochem Cytochem. 1993;41:1599–1604. doi: 10.1177/41.11.7691930. [DOI] [PubMed] [Google Scholar]
- 21.Qureshi S J, Ross A R, Ma K, Cooke H J, McIntyre M A, Chandley A C, Hargreave T B. Mol Hum Reprod. 1996;2:775–779. doi: 10.1093/molehr/2.10.775. [DOI] [PubMed] [Google Scholar]
- 22.Soulard M, Della Valle V, Siomi M C, Pinol-Roma S, Codognol P, Bauvy C, Bellini M, Lacroix J-C, Dreyfuss G, Larsen C-J. Nucleic Acids Res. 1993;21:4210–4217. doi: 10.1093/nar/21.18.4210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Le Coniat M, Soulard M, Della Valle V, Larsen C-J, Berger R. Hum Genet. 1992;88:593–595. doi: 10.1007/BF00219352. [DOI] [PubMed] [Google Scholar]
- 24.Pettersson I, Hinterberger M, Mimori T, Gottleib E, Steitz J A. J Biol Chem. 1984;259:5907–5914. [PubMed] [Google Scholar]
- 25.Lerner E A, Lerner M R, Janeway C A, Steitz J A. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Monesi V. J Cell Biol. 1964;22:521–532. doi: 10.1083/jcb.22.3.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Monesi V. Exp Cell Res. 1965;39:197–224. doi: 10.1016/0014-4827(65)90023-6. [DOI] [PubMed] [Google Scholar]
- 28.Blendy J A, Kaestner K H, Weinbauer G F, Nieschlag E, Schutz G. Nature (London) 1996;380:162–165. doi: 10.1038/380162a0. [DOI] [PubMed] [Google Scholar]
- 29.Nantel F, Monaco L, Foulkes N S, Masquilier D, Lemeur M, Henriksen K, Dierich A, Parvinen M, Sassone-Corsi P. Nature (London) 1996;380:159–162. doi: 10.1038/380159a0. [DOI] [PubMed] [Google Scholar]
- 30.Monaco L, Nantel F, Foulkes N S, Sassone-Corsi P. In: CREM: A Transcriptional Master Switch Governing the cAMP Response in the Testis. Hansson V, Levy F O, Tasken K, editors. Berlin: Springer; 1996. pp. 69–93. [Google Scholar]
- 31.Kohler M R, Vogt P H. Chromosoma. 1994;103:324–330. doi: 10.1007/BF00417879. [DOI] [PubMed] [Google Scholar]
- 32.Chandley A C, Gosden J R, Hargreave T B, Spowart G, Speed R M, McBeath S. J Med Genet. 1989;26:145–153. doi: 10.1136/jmg.26.3.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Johnsen S G. Hormones. 1970;1:1–24. doi: 10.1159/000178170. [DOI] [PubMed] [Google Scholar]
- 34.Prosser J, Inglis J D, Condie A, Ma K, Kerr S, Thakrar R, Taylor K, Cameron J M, Cooke H J. Mamm Genome. 1996;7:835–842. doi: 10.1007/s003359900246. [DOI] [PubMed] [Google Scholar]
- 35.Bedell M A, Jenkins N A, Copeland N G. Nat Genet. 1996;12:229–231. doi: 10.1038/ng0396-229. [DOI] [PubMed] [Google Scholar]
- 36.Eberhardt C G, Maines J Z, Wasserman S A. Nature (London) 1996;381:783–785. doi: 10.1038/381783a0. [DOI] [PubMed] [Google Scholar]
- 37.Kamma H, Portman D S, Dreyfuss G. Exp Cell Res. 1995;221:187–196. doi: 10.1006/excr.1995.1366. [DOI] [PubMed] [Google Scholar]