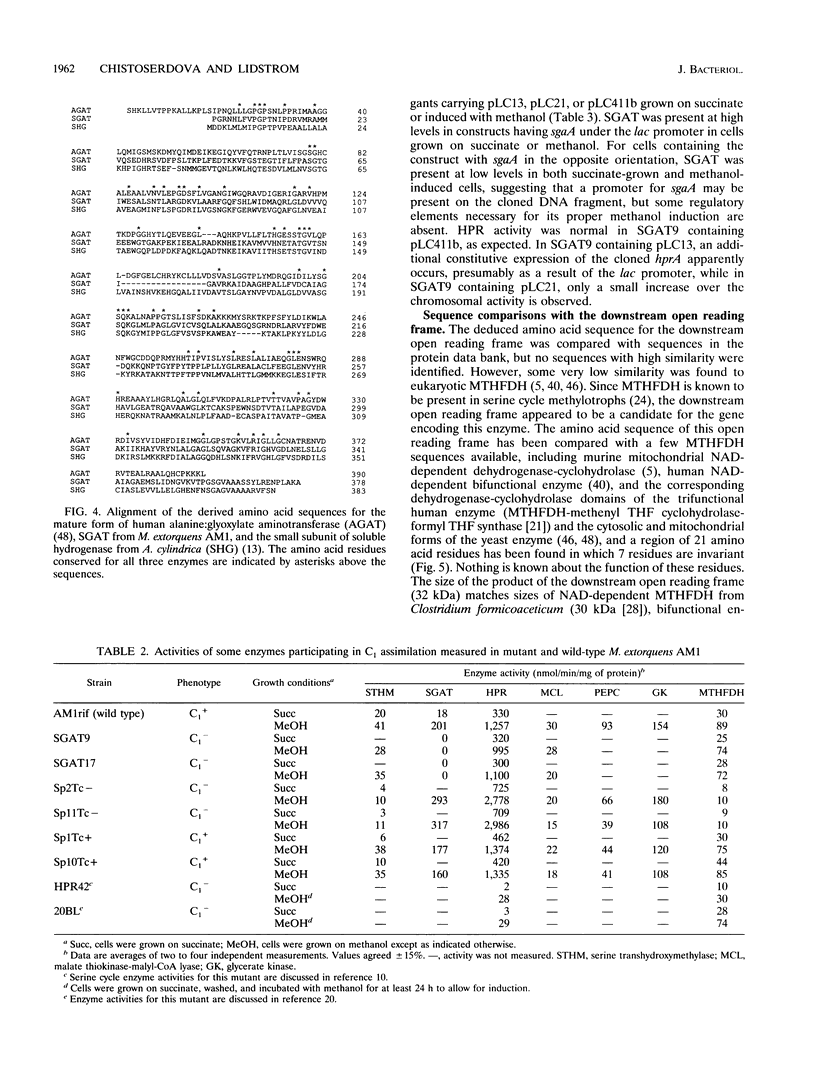
Abstract
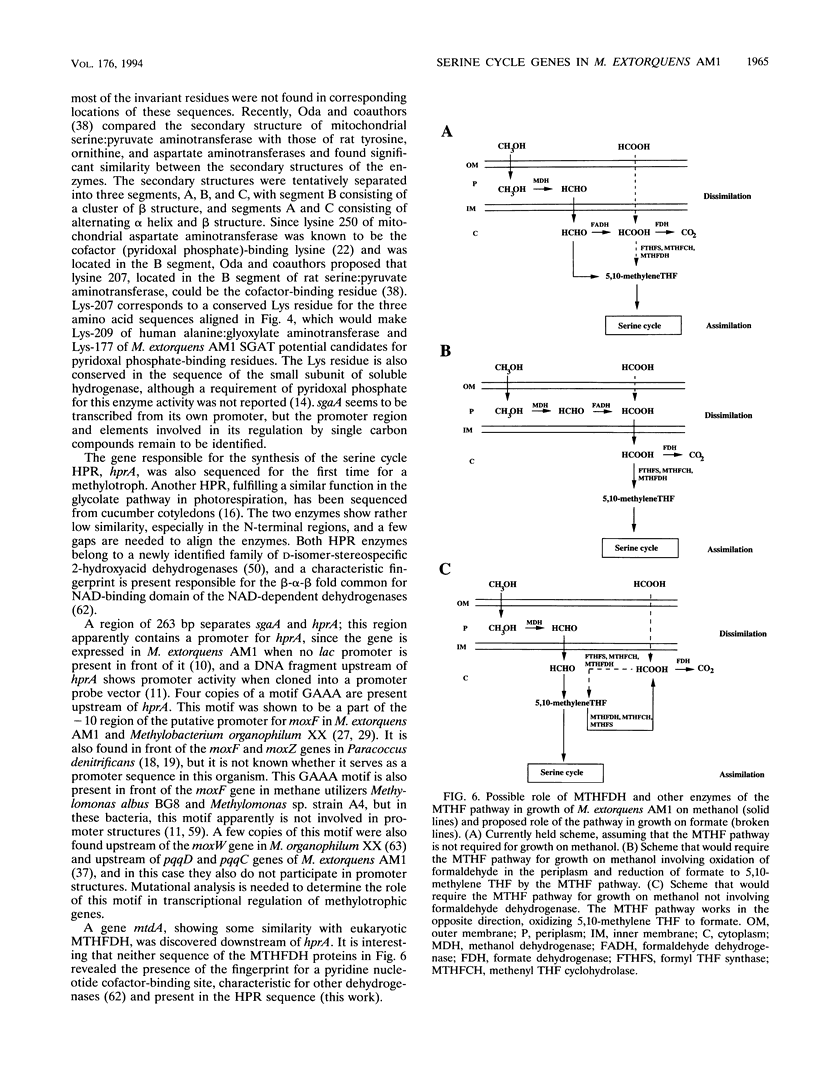
In a previous paper, we reported identification of the 5' part of hprA of Methylobacterium extorquens AM1, which encodes the serine cycle enzyme hydroxypyruvate reductase (L. V. Chistoserdova and M. E. Lidstrom, J. Bacteriol. 174:71-77, 1992). Here we present the complete sequence of hprA and partial sequence of genes adjacent to hprA. Upstream of hprA, the 3' part of an open reading frame was discovered, separated from hprA by 263 bp. This open reading frame was identified as the gene encoding another serine cycle enzyme, serine glyoxylate aminotransferase (sgaA). Cells containing an insertion mutation into sgaA were unable to grow on C1 compounds, demonstrating that the gene is required for C1 metabolism. Sequencing downstream of hprA has revealed the presence of another open reading frame (mtdA), which is probably cotranscribed with hprA. This open reading frame was identified as the gene required for the synthesis of 5,10-methylenetetrahydrofolate dehydrogenase. Our data suggest that this enzyme plays an integral role in methylotrophic metabolism in M. extorquens AM1, either in formaldehyde oxidation or as part of the serine cycle.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alfano J. R., Kahn M. L. Isolation and characterization of a gene coding for a novel aspartate aminotransferase from Rhizobium meliloti. J Bacteriol. 1993 Jul;175(13):4186–4196. doi: 10.1128/jb.175.13.4186-4196.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arps P. J., Fulton G. F., Minnich E. C., Lidstrom M. E. Genetics of serine pathway enzymes in Methylobacterium extorquens AM1: phosphoenolpyruvate carboxylase and malyl coenzyme A lyase. J Bacteriol. 1993 Jun;175(12):3776–3783. doi: 10.1128/jb.175.12.3776-3783.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur M., Molinas C., Dutka-Malen S., Courvalin P. Structural relationship between the vancomycin resistance protein VanH and 2-hydroxycarboxylic acid dehydrogenases. Gene. 1991 Jul 15;103(1):133–134. doi: 10.1016/0378-1119(91)90405-z. [DOI] [PubMed] [Google Scholar]
- Bernard N., Ferain T., Garmyn D., Hols P., Delcour J. Cloning of the D-lactate dehydrogenase gene from Lactobacillus delbrueckii subsp. bulgaricus by complementation in Escherichia coli. FEBS Lett. 1991 Sep 23;290(1-2):61–64. doi: 10.1016/0014-5793(91)81226-x. [DOI] [PubMed] [Google Scholar]
- Bélanger C., MacKenzie R. E. Isolation and characterization of cDNA clones encoding the murine NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase. J Biol Chem. 1989 Mar 25;264(9):4837–4843. [PubMed] [Google Scholar]
- Chistoserdov A. Y., Tsygankov Y. D., Lidstrom M. E. Genetic organization of methylamine utilization genes from Methylobacterium extorquens AM1. J Bacteriol. 1991 Sep;173(18):5901–5908. doi: 10.1128/jb.173.18.5901-5908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. V., Lidstrom M. E. Cloning, mutagenesis, and physiological effect of a hydroxypyruvate reductase gene from Methylobacterium extorquens AM1. J Bacteriol. 1992 Jan;174(1):71–77. doi: 10.1128/jb.174.1.71-77.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdova L. V., Lidstrom M. E. Purification and characterization of hydroxypyruvate reductase from the facultative methylotroph Methylobacterium extorquens AM1. J Bacteriol. 1991 Nov;173(22):7228–7232. doi: 10.1128/jb.173.22.7228-7232.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Dunstan P. M., Anthony C., Drabble W. T. Microbial metabolism of C 1 and C 2 compounds. The involvement of glycollate in the metabolism of ethanol and of acetate by Pseudomonas AM1. Biochem J. 1972 Jun;128(1):99–106. doi: 10.1042/bj1280099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewart G. D., Reed K. C., Smith G. D. Soluble hydrogenase of Anabaena cylindrica. Cloning and sequencing of a potential gene encoding the tritium exchange subunit. Eur J Biochem. 1990 Jan 12;187(1):215–223. doi: 10.1111/j.1432-1033.1990.tb15297.x. [DOI] [PubMed] [Google Scholar]
- Fulton G. L., Nunn D. N., Lidstrom M. E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984 Nov;160(2):718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenler J. M., Sloan J. S., Schwartz B. W., Becker W. M. Isolation, characterization and sequence analysis of a full-length cDNA clone encoding NADH-dependent hydroxypyruvate reductase from cucumber. Plant Mol Biol. 1989 Aug;13(2):139–150. doi: 10.1007/BF00016133. [DOI] [PubMed] [Google Scholar]
- Harms N., Reijnders W. N., Anazawa H., van der Palen C. J., van Spanning R. J., Oltmann L. F., Stouthamer A. H. Identification of a two-component regulatory system controlling methanol dehydrogenase synthesis in Paracoccus denitrificans. Mol Microbiol. 1993 May;8(3):457–470. doi: 10.1111/j.1365-2958.1993.tb01590.x. [DOI] [PubMed] [Google Scholar]
- Harms N., de Vries G. E., Maurer K., Hoogendijk J., Stouthamer A. H. Isolation and nucleotide sequence of the methanol dehydrogenase structural gene from Paracoccus denitrificans. J Bacteriol. 1987 Sep;169(9):3969–3975. doi: 10.1128/jb.169.9.3969-3975.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hum D. W., Bell A. W., Rozen R., MacKenzie R. E. Primary structure of a human trifunctional enzyme. Isolation of a cDNA encoding methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase. J Biol Chem. 1988 Nov 5;263(31):15946–15950. [PubMed] [Google Scholar]
- Huynh Q. K., Sakakibara R., Watanabe T., Wada M. Primary structure of mitochondrial glutamic oxaloacetic transaminase from rat liver : comparison with that of the pig heart isozyme. Biochem Biophys Res Commun. 1980 Nov 28;97(2):474–479. doi: 10.1016/0006-291x(80)90287-9. [DOI] [PubMed] [Google Scholar]
- Kalb V. F., Jr, Bernlohr R. W. A new spectrophotometric assay for protein in cell extracts. Anal Biochem. 1977 Oct;82(2):362–371. doi: 10.1016/0003-2697(77)90173-7. [DOI] [PubMed] [Google Scholar]
- Large P. J., Quayle J. R. Microbial growth on C(1) compounds. 5. Enzyme activities in extracts of Pseudomonas AM1. Biochem J. 1963 May;87(2):386–396. doi: 10.1042/bj0870386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerch H. P., Blöcker H., Kallwass H., Hoppe J., Tsai H., Collins J. Cloning, sequencing and expression in Escherichia coli of the D-2-hydroxyisocaproate dehydrogenase gene of Lactobacillus casei. Gene. 1989 May 15;78(1):47–57. doi: 10.1016/0378-1119(89)90313-2. [DOI] [PubMed] [Google Scholar]
- Ljungdahl L. G., O'Brien W. E., Moore M. R., Liu M. T. Methylenetetrahydrofolate dehydrogenase from Clostridium formicoaceticum and methylenetetrahydrofolate dehydrogenase, methenyltetrahydrofolate cyclohydrolase (combined) from Clostridium thermoaceticum. Methods Enzymol. 1980;66:599–609. doi: 10.1016/0076-6879(80)66513-6. [DOI] [PubMed] [Google Scholar]
- Machlin S. M., Hanson R. S. Nucleotide sequence and transcriptional start site of the Methylobacterium organophilum XX methanol dehydrogenase structural gene. J Bacteriol. 1988 Oct;170(10):4739–4747. doi: 10.1128/jb.170.10.4739-4747.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta P. K., Hale T. I., Christen P. Evolutionary relationships among aminotransferases. Tyrosine aminotransferase, histidinol-phosphate aminotransferase, and aspartate aminotransferase are homologous proteins. Eur J Biochem. 1989 Dec 8;186(1-2):249–253. doi: 10.1111/j.1432-1033.1989.tb15202.x. [DOI] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Mejia N. R., Rios-Orlandi E. M., MacKenzie R. E. NAD-dependent methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase from ascites tumor cells. Purification and properties. J Biol Chem. 1986 Jul 15;261(20):9509–9513. [PubMed] [Google Scholar]
- Miyata A., Yoshida T., Yamaguchi K., Yokoyama C., Tanabe T., Toh H., Mitsunaga T., Izumi Y. Molecular cloning and expression of the gene for serine hydroxymethyltransferase from an obligate methylotroph Hyphomicrobium methylovorum GM2. Eur J Biochem. 1993 Mar 15;212(3):745–750. doi: 10.1111/j.1432-1033.1993.tb17713.x. [DOI] [PubMed] [Google Scholar]
- Morris C. J., Biville F., Turlin E., Lee E., Ellermann K., Fan W. H., Ramamoorthi R., Springer A. L., Lidstrom M. E. Isolation, phenotypic characterization, and complementation analysis of mutants of Methylobacterium extorquens AM1 unable to synthesize pyrroloquinoline quinone and sequences of pqqD, pqqG, and pqqC. J Bacteriol. 1994 Mar;176(6):1746–1755. doi: 10.1128/jb.176.6.1746-1755.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oda T., Miyajima H., Suzuki Y., Ichiyama A. Nucleotide sequence of the cDNA encoding the precursor for mitochondrial serine:pyruvate aminotransferase of rat liver. Eur J Biochem. 1987 Nov 2;168(3):537–542. doi: 10.1111/j.1432-1033.1987.tb13451.x. [DOI] [PubMed] [Google Scholar]
- PEEL D., QUAYLE J. R. Microbial growth on C1 compounds. I. Isolation and characterization of Pseudomonas AM 1. Biochem J. 1961 Dec;81:465–469. doi: 10.1042/bj0810465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peri K. G., Belanger C., Mackenzie R. E. Nucleotide sequence of the human NAD-dependent methylene tetrahydrofolate dehydrogenase-cyclohydrolase. Nucleic Acids Res. 1989 Nov 11;17(21):8853–8853. doi: 10.1093/nar/17.21.8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Popov V. O., Shumilin I. A., Ustinnikova T. B., Lamzin V. S., Egorov Ts A. NAD-zavisimaia formiatdegidrogenaza metilotrofnykh bakterii Pseudomonas sp. 101. I. Aminokislotnaia posledovatel'nost'. Bioorg Khim. 1990 Mar;16(3):324–335. [PubMed] [Google Scholar]
- Purdue P. E., Takada Y., Danpure C. J. Identification of mutations associated with peroxisome-to-mitochondrion mistargeting of alanine/glyoxylate aminotransferase in primary hyperoxaluria type 1. J Cell Biol. 1990 Dec;111(6 Pt 1):2341–2351. doi: 10.1083/jcb.111.6.2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Shannon K. W., Rabinowitz J. C. Isolation and characterization of the Saccharomyces cerevisiae MIS1 gene encoding mitochondrial C1-tetrahydrofolate synthase. J Biol Chem. 1988 Jun 5;263(16):7717–7725. [PubMed] [Google Scholar]
- Staben C., Rabinowitz J. C. Nucleotide sequence of the Saccharomyces cerevisiae ADE3 gene encoding C1-tetrahydrofolate synthase. J Biol Chem. 1986 Apr 5;261(10):4629–4637. [PubMed] [Google Scholar]
- Taguchi H., Ohta T. D-lactate dehydrogenase is a member of the D-isomer-specific 2-hydroxyacid dehydrogenase family. Cloning, sequencing, and expression in Escherichia coli of the D-lactate dehydrogenase gene of Lactobacillus plantarum. J Biol Chem. 1991 Jul 5;266(19):12588–12594. [PubMed] [Google Scholar]
- Takada Y., Kaneko N., Esumi H., Purdue P. E., Danpure C. J. Human peroxisomal L-alanine: glyoxylate aminotransferase. Evolutionary loss of a mitochondrial targeting signal by point mutation of the initiation codon. Biochem J. 1990 Jun 1;268(2):517–520. doi: 10.1042/bj2680517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan L. U., MacKenzie R. E. Methylenetetrahydrofolate dehydrogenase-methenyltetrahydrofolate cyclohydrolase-formyltetrahydrofolate synthetase from porcine liver. Location of the activities in two domains of the multifunctional polypeptide. Can J Biochem. 1979 Jun;57(6):806–812. doi: 10.1139/o79-100. [DOI] [PubMed] [Google Scholar]
- Tobey K. L., Grant G. A. The nucleotide sequence of the serA gene of Escherichia coli and the amino acid sequence of the encoded protein, D-3-phosphoglycerate dehydrogenase. J Biol Chem. 1986 Sep 15;261(26):12179–12183. [PubMed] [Google Scholar]
- Tolbert N. E. Metabolic pathways in peroxisomes and glyoxysomes. Annu Rev Biochem. 1981;50:133–157. doi: 10.1146/annurev.bi.50.070181.001025. [DOI] [PubMed] [Google Scholar]
- Tsuji K., Tsien H. C., Hanson R. S., DePalma S. R., Scholtz R., LaRoche S. 16S ribosomal RNA sequence analysis for determination of phylogenetic relationship among methylotrophs. J Gen Microbiol. 1990 Jan;136(1):1–10. doi: 10.1099/00221287-136-1-1. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. The pUC plasmids, an M13mp7-derived system for insertion mutagenesis and sequencing with synthetic universal primers. Gene. 1982 Oct;19(3):259–268. doi: 10.1016/0378-1119(82)90015-4. [DOI] [PubMed] [Google Scholar]
- Watson R. J., Rastogi V. K. Cloning and nucleotide sequencing of Rhizobium meliloti aminotransferase genes: an aspartate aminotransferase required for symbiotic nitrogen fixation is atypical. J Bacteriol. 1993 Apr;175(7):1919–1928. doi: 10.1128/jb.175.7.1919-1928.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker J. R., Granum P. E. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal Biochem. 1980 Nov 15;109(1):156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]
- Wierenga R. K., Terpstra P., Hol W. G. Prediction of the occurrence of the ADP-binding beta alpha beta-fold in proteins, using an amino acid sequence fingerprint. J Mol Biol. 1986 Jan 5;187(1):101–107. doi: 10.1016/0022-2836(86)90409-2. [DOI] [PubMed] [Google Scholar]
- Xu H. H., Viebahn M., Hanson R. S. Identification of methanol-regulated promoter sequences from the facultative methylotrophic bacterium Methylobacterium organophilum XX. J Gen Microbiol. 1993 Apr;139(4):743–752. doi: 10.1099/00221287-139-4-743. [DOI] [PubMed] [Google Scholar]
- van der Oost J., van Walraven H. S., Bogerd J., Smit A. B., Ewart G. D., Smith G. D. Nucleotide sequence of the gene proposed to encode the small subunit of the soluble hydrogenase of the thermophilic unicellular cyanobacterium Synechococcus PCC 6716. Nucleic Acids Res. 1989 Dec 11;17(23):10098–10098. [PMC free article] [PubMed] [Google Scholar]



