Abstract
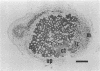
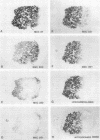
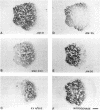
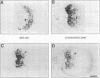
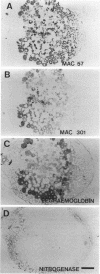
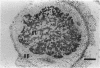
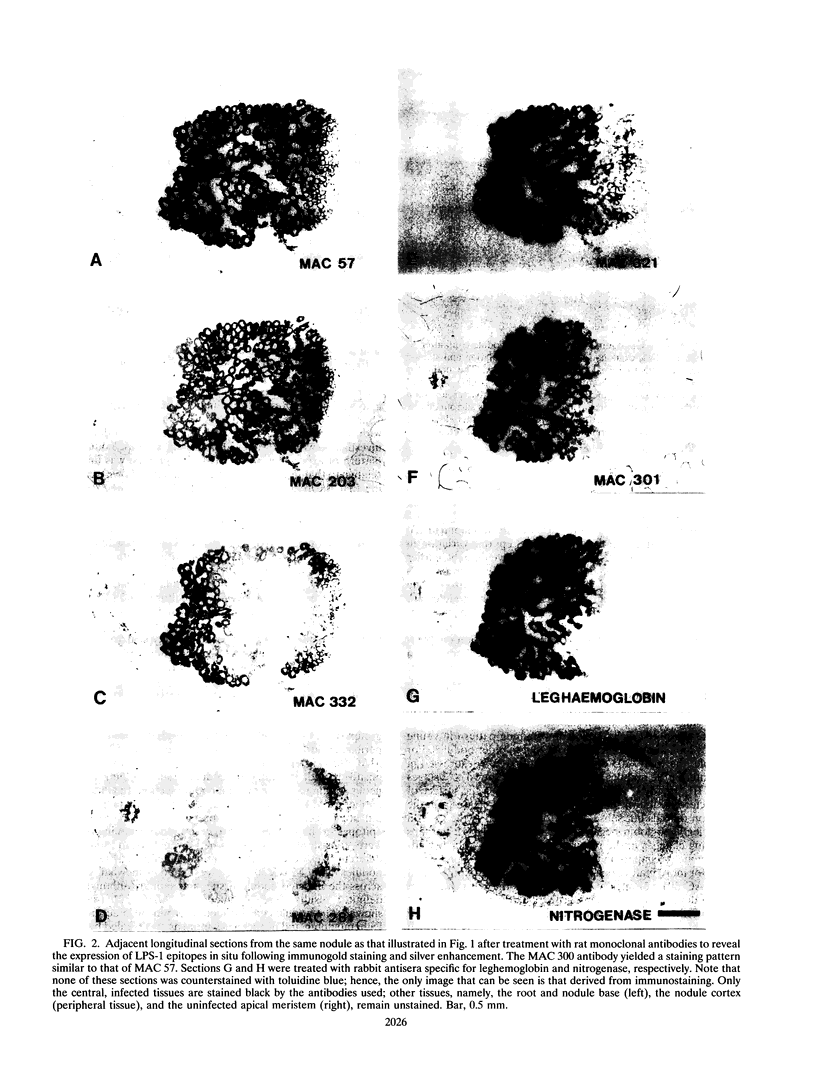
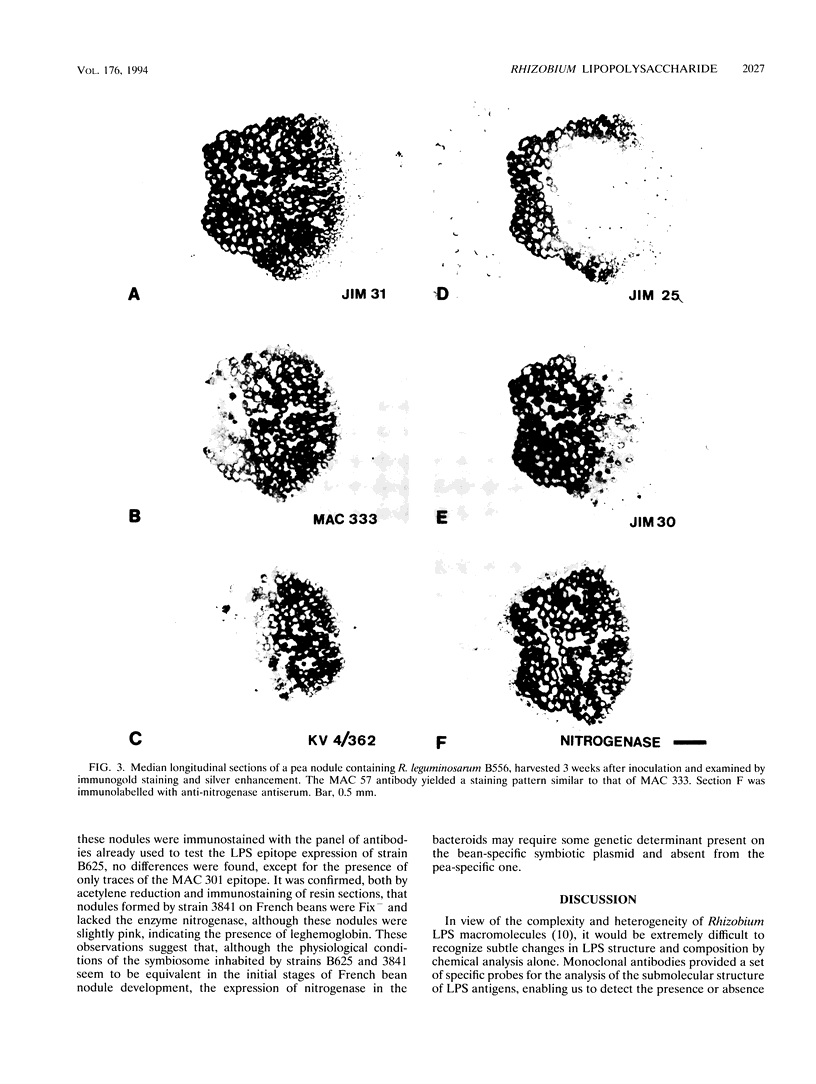
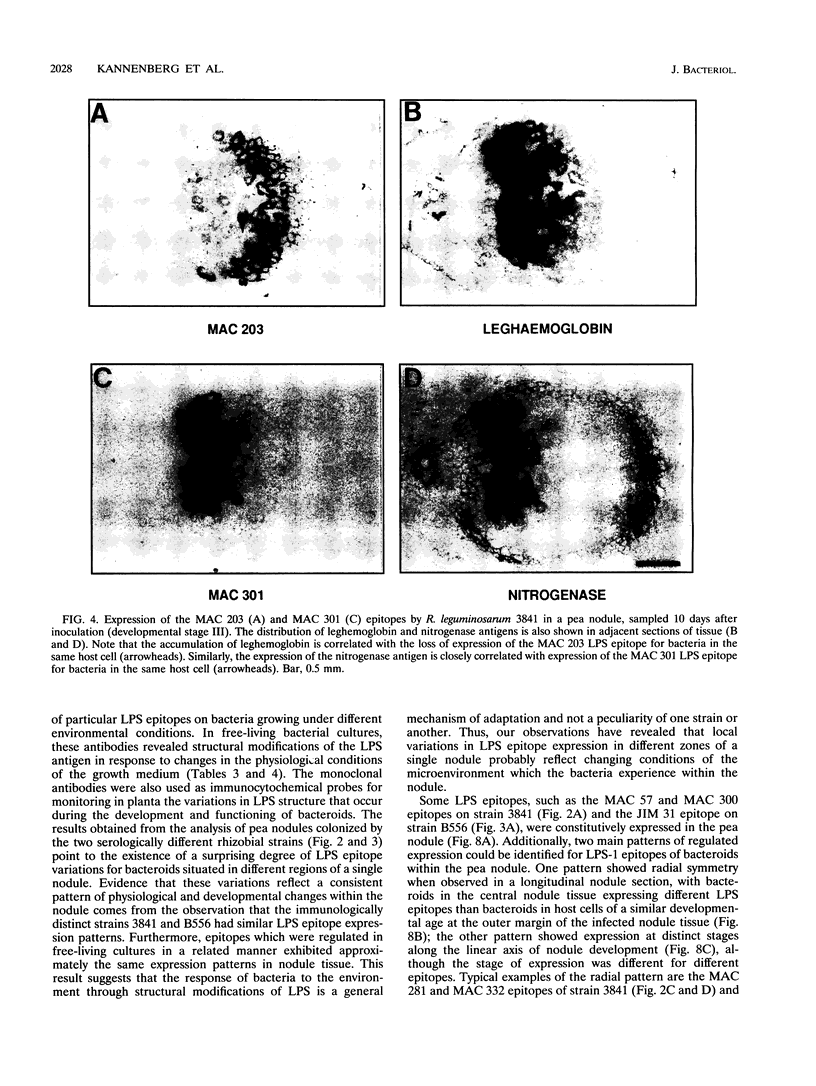
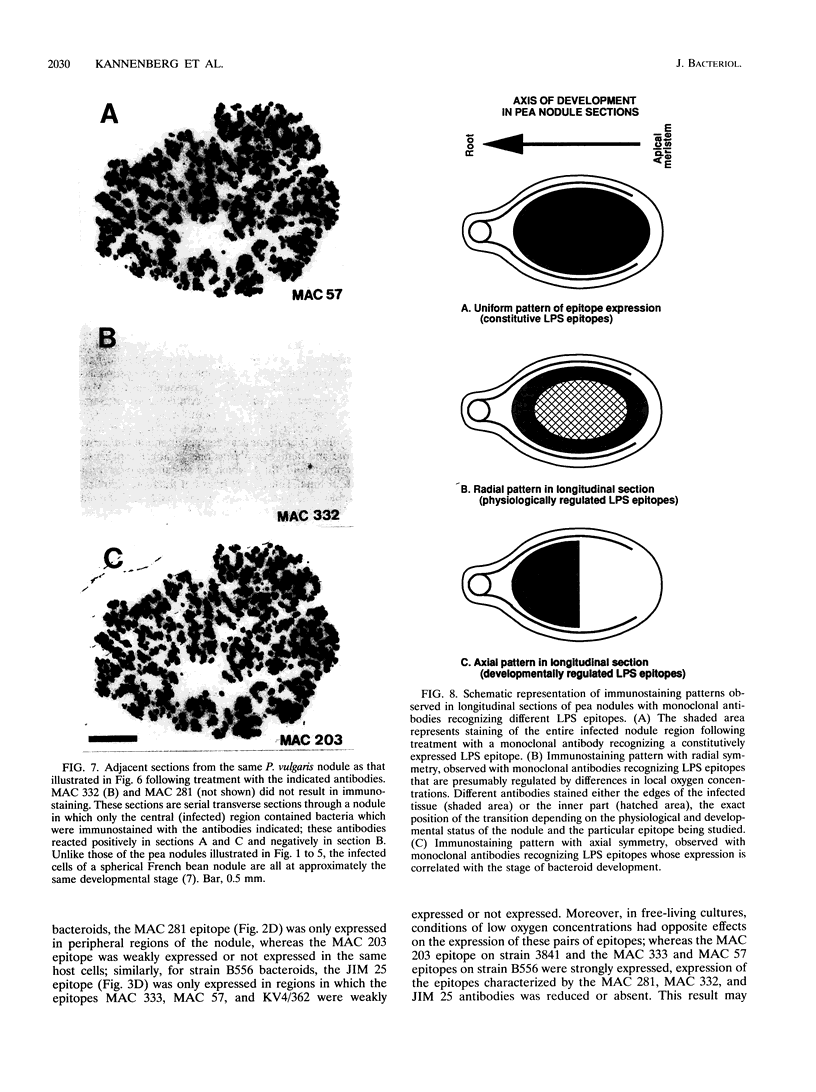
To investigate the in situ expression of lipopolysaccharide (LPS) epitopes on nodule bacteria of Rhizobium leguminosarum, monoclonal antibodies recognizing LPS macromolecules were used for immunocytochemical staining of pea nodule tissue. Many LPS epitopes were constitutively expressed, and the corresponding antibodies reacted in nodule sections with bacteria at all stages of tissue infection and cell invasion. Some antibodies, however, recognized epitopes that were only expressed in particular regions of the nodule. Two general patterns of regulated LPS epitope expression could be distinguished on longitudinal sections of nodules. A radial pattern probably reflected the local physiological conditions experienced by endosymbiotic bacteria as a result of oxygen diffusion into the nodule tissue. The other pattern of expression, which followed a linear axis of symmetry along a longitudinal section of the pea nodule, was apparently associated with the differentiation of nodule bacteria and the development of the nitrogen-fixing capacity in bacteroids. Basically similar patterns of LPS epitope expression were observed for pea nodules harboring either of two immunologically distinct strains of R. leguminosarum bv. viciae, although these epitopes were recognized by different sets of strain-specific monoclonal antibodies. Furthermore, LPS epitope expression of rhizobia in pea nodules was compared with that of equivalent strains in nodules of French bean (Phaseolus vulgaris). From these observations, it is suggested that structural modifications of Rhizobium LPS may play an important role in the adaptation of endosymbiotic rhizobia to the surrounding microenvironment.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bhat U. R., Carlson R. W. Chemical characterization of pH-dependent structural epitopes of lipopolysaccharides from Rhizobium leguminosarum biovar phaseoli. J Bacteriol. 1992 Apr;174(7):2230–2235. doi: 10.1128/jb.174.7.2230-2235.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brewin N. J. Development of the legume root nodule. Annu Rev Cell Biol. 1991;7:191–226. doi: 10.1146/annurev.cb.07.110191.001203. [DOI] [PubMed] [Google Scholar]
- Brewin N. J., Robertson J. G., Wood E. A., Wells B., Larkins A. P., Galfre G., Butcher G. W. Monoclonal antibodies to antigens in the peribacteroid membrane from Rhizobium-induced root nodules of pea cross-react with plasma membranes and Golgi bodies. EMBO J. 1985 Mar;4(3):605–611. doi: 10.1002/j.1460-2075.1985.tb03673.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hancock R. E. Alterations in outer membrane permeability. Annu Rev Microbiol. 1984;38:237–264. doi: 10.1146/annurev.mi.38.100184.001321. [DOI] [PubMed] [Google Scholar]
- Hennecke H. Nitrogen fixation genes involved in the Bradyrhizobium japonicum-soybean symbiosis. FEBS Lett. 1990 Aug 1;268(2):422–426. doi: 10.1016/0014-5793(90)81297-2. [DOI] [PubMed] [Google Scholar]
- Johnston A. W., Beringer J. E. Identification of the rhizobium strains in pea root nodules using genetic markers. J Gen Microbiol. 1975 Apr;87(2):343–350. doi: 10.1099/00221287-87-2-343. [DOI] [PubMed] [Google Scholar]
- Kannenberg E. L., Brewin N. J. Expression of a cell surface antigen from Rhizobium leguminosarum 3841 is regulated by oxygen and pH. J Bacteriol. 1989 Sep;171(9):4543–4548. doi: 10.1128/jb.171.9.4543-4548.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kannenberg E. L., Rathbun E. A., Brewin N. J. Molecular dissection of structure and function in the lipopolysaccharide of Rhizobium leguminosarum strain 3841 using monoclonal antibodies and genetic analysis. Mol Microbiol. 1992 Sep;6(17):2477–2487. doi: 10.1111/j.1365-2958.1992.tb01424.x. [DOI] [PubMed] [Google Scholar]
- Kneen B. E., Larue T. A., Hirsch A. M., Smith C. A., Weeden N. F. sym 13-A Gene Conditioning Ineffective Nodulation in Pisum sativum. Plant Physiol. 1990 Nov;94(3):899–905. doi: 10.1104/pp.94.3.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noel K. D., Vandenbosch K. A., Kulpaca B. Mutations in Rhizobium phaseoli that lead to arrested development of infection threads. J Bacteriol. 1986 Dec;168(3):1392–1401. doi: 10.1128/jb.168.3.1392-1401.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priefer U. B. Genes involved in lipopolysaccharide production and symbiosis are clustered on the chromosome of Rhizobium leguminosarum biovar viciae VF39. J Bacteriol. 1989 Nov;171(11):6161–6168. doi: 10.1128/jb.171.11.6161-6168.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raetz C. R. Bacterial endotoxins: extraordinary lipids that activate eucaryotic signal transduction. J Bacteriol. 1993 Sep;175(18):5745–5753. doi: 10.1128/jb.175.18.5745-5753.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sindhu S. S., Brewin N. J., Kannenberg E. L. Immunochemical analysis of lipopolysaccharides from free-living and endosymbiotic forms of Rhizobium leguminosarum. J Bacteriol. 1990 Apr;172(4):1804–1813. doi: 10.1128/jb.172.4.1804-1813.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H., Brewin N. J., Noel K. D. Rhizobium leguminosarum CFN42 lipopolysaccharide antigenic changes induced by environmental conditions. J Bacteriol. 1992 Apr;174(7):2222–2229. doi: 10.1128/jb.174.7.2222-2229.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VandenBosch K. A., Brewin N. J., Kannenberg E. L. Developmental regulation of a Rhizobium cell surface antigen during growth of pea root nodules. J Bacteriol. 1989 Sep;171(9):4537–4542. doi: 10.1128/jb.171.9.4537-4542.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandenbosch K. A., Bradley D. J., Knox J. P., Perotto S., Butcher G. W., Brewin N. J. Common components of the infection thread matrix and the intercellular space identified by immunocytochemical analysis of pea nodules and uninfected roots. EMBO J. 1989 Feb;8(2):335–341. doi: 10.1002/j.1460-2075.1989.tb03382.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wood E. A., Butcher G. W., Brewin N. J., Kannenberg E. L. Genetic derepression of a developmentally regulated lipopolysaccharide antigen from Rhizobium leguminosarum 3841. J Bacteriol. 1989 Sep;171(9):4549–4555. doi: 10.1128/jb.171.9.4549-4555.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Maagd R. A., Rao A. S., Mulders I. H., Goosen-de Roo L., van Loosdrecht M. C., Wijffelman C. A., Lugtenberg B. J. Isolation and characterization of mutants of Rhizobium leguminosarum bv. viciae 248 with altered lipopolysaccharides: possible role of surface charge or hydrophobicity in bacterial release from the infection thread. J Bacteriol. 1989 Feb;171(2):1143–1150. doi: 10.1128/jb.171.2.1143-1150.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]