Abstract
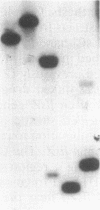
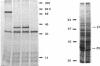
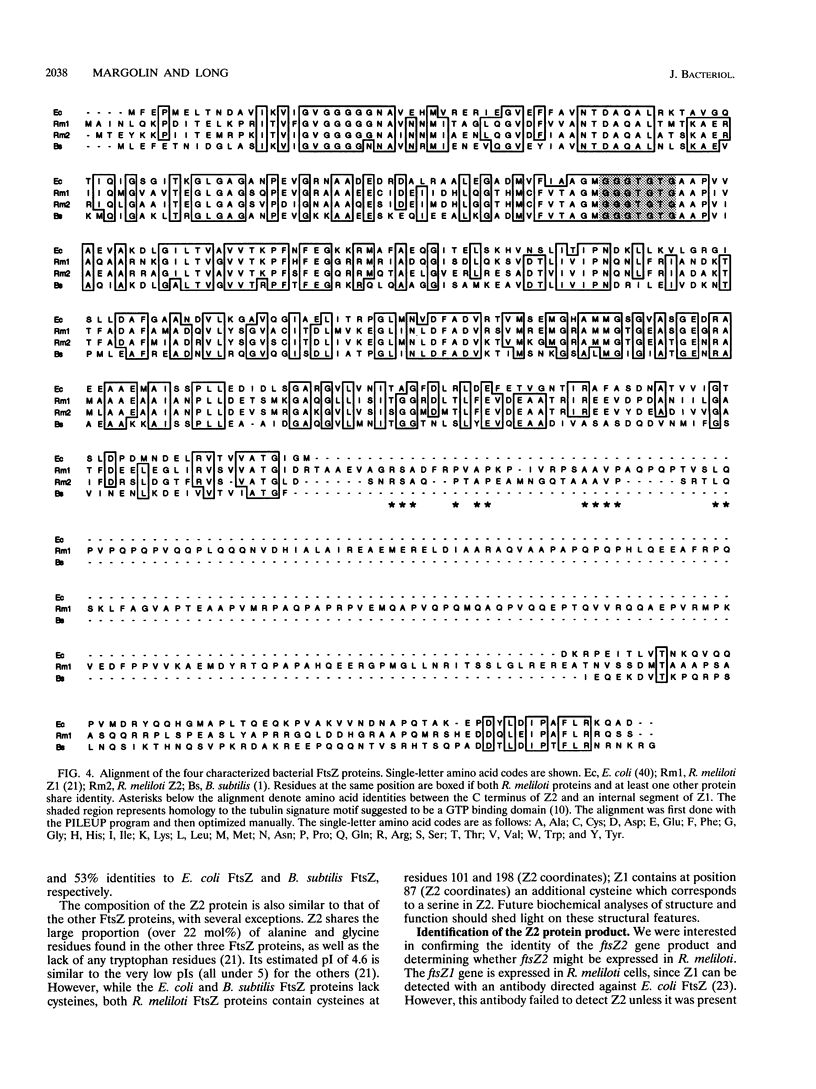
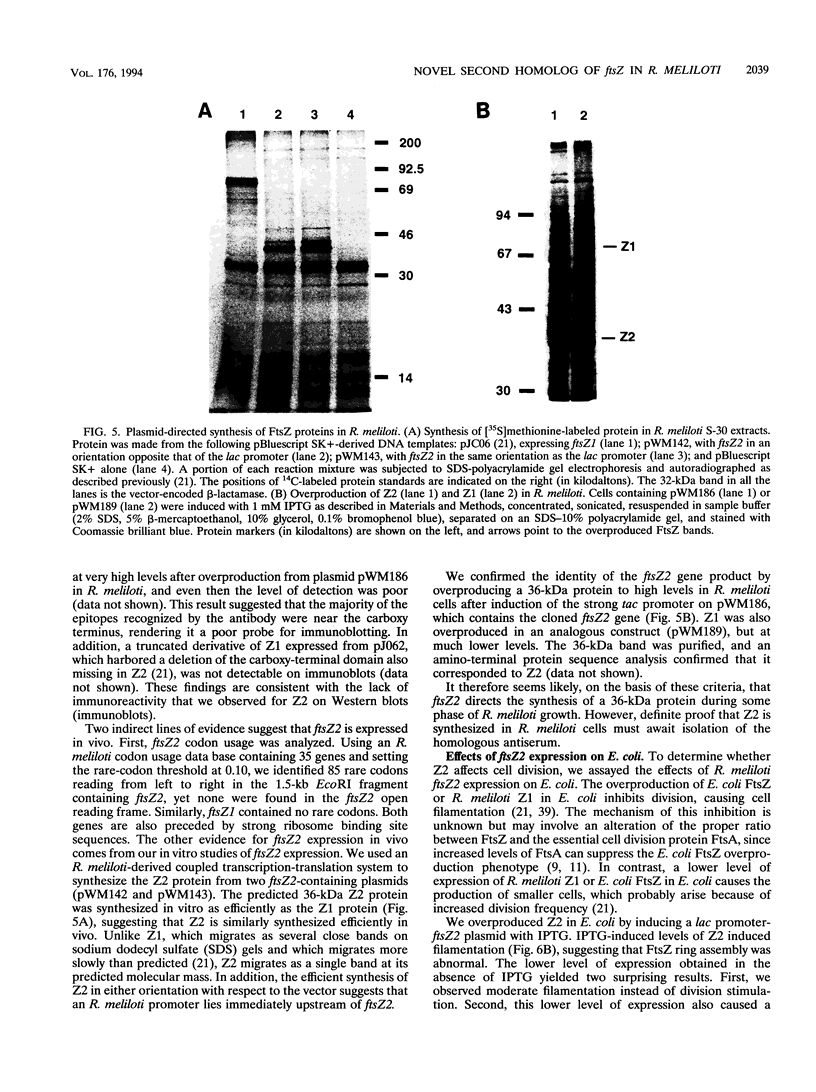
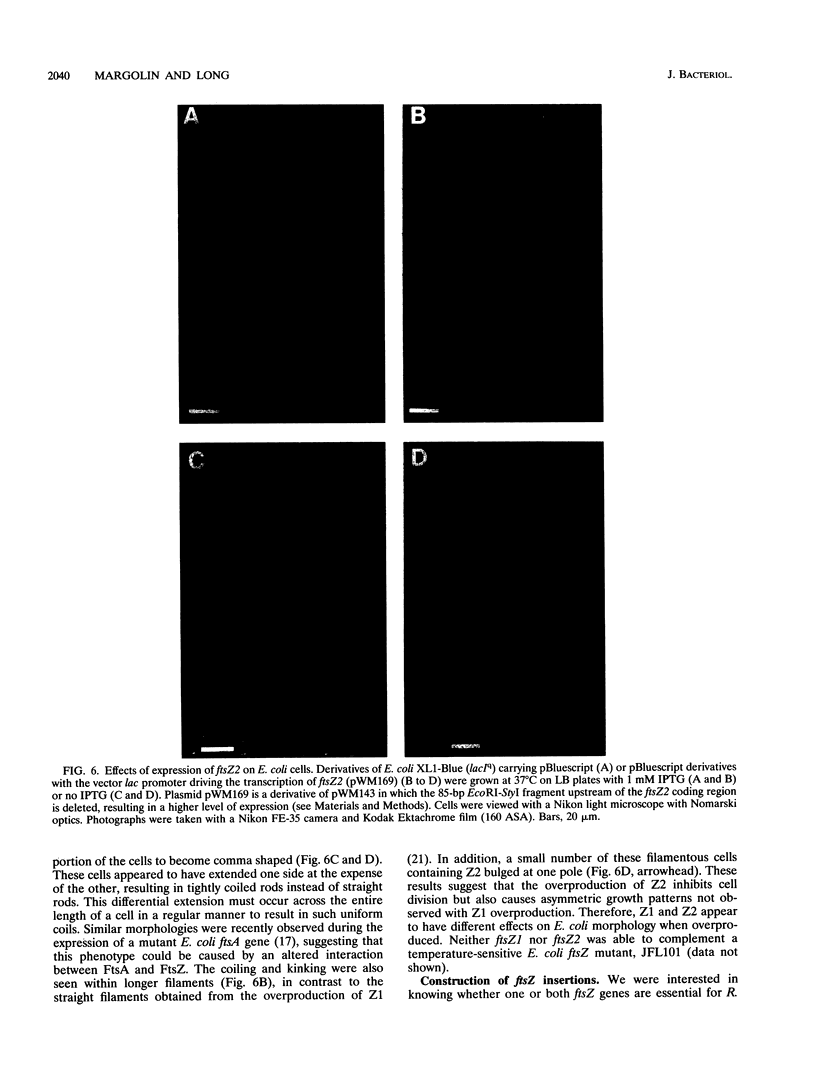
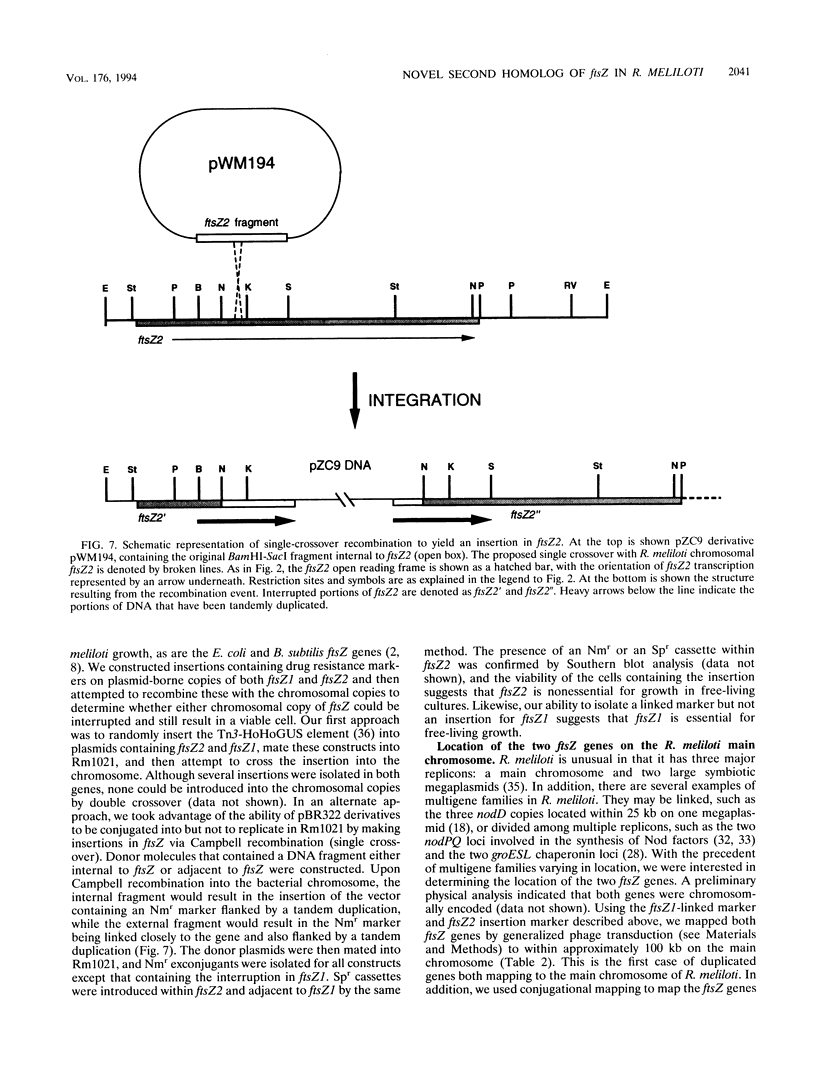
We have identified a second homolog of the cell division gene, ftsZ, in the endosymbiont Rhizobium meliloti. The ftsZ2 gene was cloned by screening a genomic lambda library with a probe derived from PCR amplification of a highly conserved domain. It encodes a 36-kDa protein which shares a high level of sequence similarity with the FtsZ proteins of Escherichia coli and Bacillus subtilis and FtsZ1 (Z1) of R. meliloti but lacks the carboxy-terminal region conserved in other FtsZ proteins. The identity of the ftsZ2 gene product was confirmed both by in vitro transcription-translation in an R. meliloti S-30 extract and by overproduction in R. meliloti cells. As with Z1, the overproduction of FtsZ2 in E. coli inhibited cell division and induced filamentation, although to a lesser extent than with Z1. However, the expression of ftsZ2 in E. coli under certain conditions caused some cells to coil dramatically, a phenotype not observed during Z1 overproduction. Although several Tn3-GUS (glucuronidase) insertions in a plasmid-borne ftsZ2 gene failed to cross into the chromosome, one interruption in the chromosomal ftsZ2 gene was isolated, suggesting that ftsZ2 is nonessential for viability. The two ftsZ genes were genetically mapped to the R. meliloti main chromosome, approximately 100 kb apart.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Beall B., Lowe M., Lutkenhaus J. Cloning and characterization of Bacillus subtilis homologs of Escherichia coli cell division genes ftsZ and ftsA. J Bacteriol. 1988 Oct;170(10):4855–4864. doi: 10.1128/jb.170.10.4855-4864.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beall B., Lutkenhaus J. FtsZ in Bacillus subtilis is required for vegetative septation and for asymmetric septation during sporulation. Genes Dev. 1991 Mar;5(3):447–455. doi: 10.1101/gad.5.3.447. [DOI] [PubMed] [Google Scholar]
- Begg K. J., Donachie W. D. Cell shape and division in Escherichia coli: experiments with shape and division mutants. J Bacteriol. 1985 Aug;163(2):615–622. doi: 10.1128/jb.163.2.615-622.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bi E. F., Lutkenhaus J. FtsZ ring structure associated with division in Escherichia coli. Nature. 1991 Nov 14;354(6349):161–164. doi: 10.1038/354161a0. [DOI] [PubMed] [Google Scholar]
- Bi E., Lutkenhaus J. Analysis of ftsZ mutations that confer resistance to the cell division inhibitor SulA (SfiA). J Bacteriol. 1990 Oct;172(10):5602–5609. doi: 10.1128/jb.172.10.5602-5609.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C. Y., Winans S. C. Controlled expression of the transcriptional activator gene virG in Agrobacterium tumefaciens by using the Escherichia coli lac promoter. J Bacteriol. 1991 Feb;173(3):1139–1144. doi: 10.1128/jb.173.3.1139-1144.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Church G. M., Gilbert W. Genomic sequencing. Proc Natl Acad Sci U S A. 1984 Apr;81(7):1991–1995. doi: 10.1073/pnas.81.7.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K., Lutkenhaus J. The proper ratio of FtsZ to FtsA is required for cell division to occur in Escherichia coli. J Bacteriol. 1992 Oct;174(19):6145–6151. doi: 10.1128/jb.174.19.6145-6151.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai K., Lutkenhaus J. ftsZ is an essential cell division gene in Escherichia coli. J Bacteriol. 1991 Jun;173(11):3500–3506. doi: 10.1128/jb.173.11.3500-3506.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewar S. J., Begg K. J., Donachie W. D. Inhibition of cell division initiation by an imbalance in the ratio of FtsA to FtsZ. J Bacteriol. 1992 Oct;174(19):6314–6316. doi: 10.1128/jb.174.19.6314-6316.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doherty D., Leigh J. A., Glazebrook J., Walker G. C. Rhizobium meliloti mutants that overproduce the R. meliloti acidic calcofluor-binding exopolysaccharide. J Bacteriol. 1988 Sep;170(9):4249–4256. doi: 10.1128/jb.170.9.4249-4256.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Finan T. M., Kunkel B., De Vos G. F., Signer E. R. Second symbiotic megaplasmid in Rhizobium meliloti carrying exopolysaccharide and thiamine synthesis genes. J Bacteriol. 1986 Jul;167(1):66–72. doi: 10.1128/jb.167.1.66-72.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fisher R. F., Swanson J. A., Mulligan J. T., Long S. R. Extended Region of Nodulation Genes in Rhizobium meliloti 1021. II. Nucleotide Sequence, Transcription Start Sites and Protein Products. Genetics. 1987 Oct;117(2):191–201. doi: 10.1093/genetics/117.2.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gayda R. C., Henk M. C., Leong D. C-shaped cells caused by expression of an ftsA mutation in Escherichia coli. J Bacteriol. 1992 Aug;174(16):5362–5370. doi: 10.1128/jb.174.16.5362-5370.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Honma M. A., Asomaning M., Ausubel F. M. Rhizobium meliloti nodD genes mediate host-specific activation of nodABC. J Bacteriol. 1990 Feb;172(2):901–911. doi: 10.1128/jb.172.2.901-911.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein S., Lohman K., Clover R., Walker G. C., Signer E. R. A directional, high-frequency chromosomal mobilization system for genetic mapping of Rhizobium meliloti. J Bacteriol. 1992 Jan;174(1):324–326. doi: 10.1128/jb.174.1.324-326.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. R. Rhizobium-legume nodulation: life together in the underground. Cell. 1989 Jan 27;56(2):203–214. doi: 10.1016/0092-8674(89)90893-3. [DOI] [PubMed] [Google Scholar]
- Margolin W., Corbo J. C., Long S. R. Cloning and characterization of a Rhizobium meliloti homolog of the Escherichia coli cell division gene ftsZ. J Bacteriol. 1991 Sep;173(18):5822–5830. doi: 10.1128/jb.173.18.5822-5830.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin M. O., Long S. R. Generalized transduction in Rhizobium meliloti. J Bacteriol. 1984 Jul;159(1):125–129. doi: 10.1128/jb.159.1.125-129.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meade H. M., Long S. R., Ruvkun G. B., Brown S. E., Ausubel F. M. Physical and genetic characterization of symbiotic and auxotrophic mutants of Rhizobium meliloti induced by transposon Tn5 mutagenesis. J Bacteriol. 1982 Jan;149(1):114–122. doi: 10.1128/jb.149.1.114-122.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukherjee A., Dai K., Lutkenhaus J. Escherichia coli cell division protein FtsZ is a guanine nucleotide binding protein. Proc Natl Acad Sci U S A. 1993 Feb 1;90(3):1053–1057. doi: 10.1073/pnas.90.3.1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulligan J. T., Long S. R. Induction of Rhizobium meliloti nodC expression by plant exudate requires nodD. Proc Natl Acad Sci U S A. 1985 Oct;82(19):6609–6613. doi: 10.1073/pnas.82.19.6609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- RayChaudhuri D., Park J. T. Escherichia coli cell-division gene ftsZ encodes a novel GTP-binding protein. Nature. 1992 Sep 17;359(6392):251–254. doi: 10.1038/359251a0. [DOI] [PubMed] [Google Scholar]
- Schwedock J., Long S. R. ATP sulphurylase activity of the nodP and nodQ gene products of Rhizobium meliloti. Nature. 1990 Dec 13;348(6302):644–647. doi: 10.1038/348644a0. [DOI] [PubMed] [Google Scholar]
- Schwedock J., Long S. R. Nucleotide sequence and protein products of two new nodulation genes of Rhizobium meliloti, nodP and nodQ. Mol Plant Microbe Interact. 1989 Jul-Aug;2(4):181–194. doi: 10.1094/mpmi-2-181. [DOI] [PubMed] [Google Scholar]
- Snyder J. A., McIntosh J. R. Biochemistry and physiology of microtubules. Annu Rev Biochem. 1976;45:699–720. doi: 10.1146/annurev.bi.45.070176.003411. [DOI] [PubMed] [Google Scholar]
- Sobral B. W., Honeycutt R. J., Atherly A. G., McClelland M. Electrophoretic separation of the three Rhizobium meliloti replicons. J Bacteriol. 1991 Aug;173(16):5173–5180. doi: 10.1128/jb.173.16.5173-5180.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson J. A., Mulligan J. T., Long S. R. Regulation of syrM and nodD3 in Rhizobium meliloti. Genetics. 1993 Jun;134(2):435–444. doi: 10.1093/genetics/134.2.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz D., Renz M. An optimized freeze-squeeze method for the recovery of DNA fragments from agarose gels. Anal Biochem. 1983 Jul 1;132(1):14–19. doi: 10.1016/0003-2697(83)90419-0. [DOI] [PubMed] [Google Scholar]
- Vieira J., Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- Ward J. E., Jr, Lutkenhaus J. Overproduction of FtsZ induces minicell formation in E. coli. Cell. 1985 Oct;42(3):941–949. doi: 10.1016/0092-8674(85)90290-9. [DOI] [PubMed] [Google Scholar]
- Wu T. T. A model for three-point analysis of random general transduction. Genetics. 1966 Aug;54(2):405–410. doi: 10.1093/genetics/54.2.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi Q. M., Rockenbach S., Ward J. E., Jr, Lutkenhaus J. Structure and expression of the cell division genes ftsQ, ftsA and ftsZ. J Mol Biol. 1985 Aug 5;184(3):399–412. doi: 10.1016/0022-2836(85)90290-6. [DOI] [PubMed] [Google Scholar]
- de Boer P., Crossley R., Rothfield L. The essential bacterial cell-division protein FtsZ is a GTPase. Nature. 1992 Sep 17;359(6392):254–256. doi: 10.1038/359254a0. [DOI] [PubMed] [Google Scholar]