Abstract
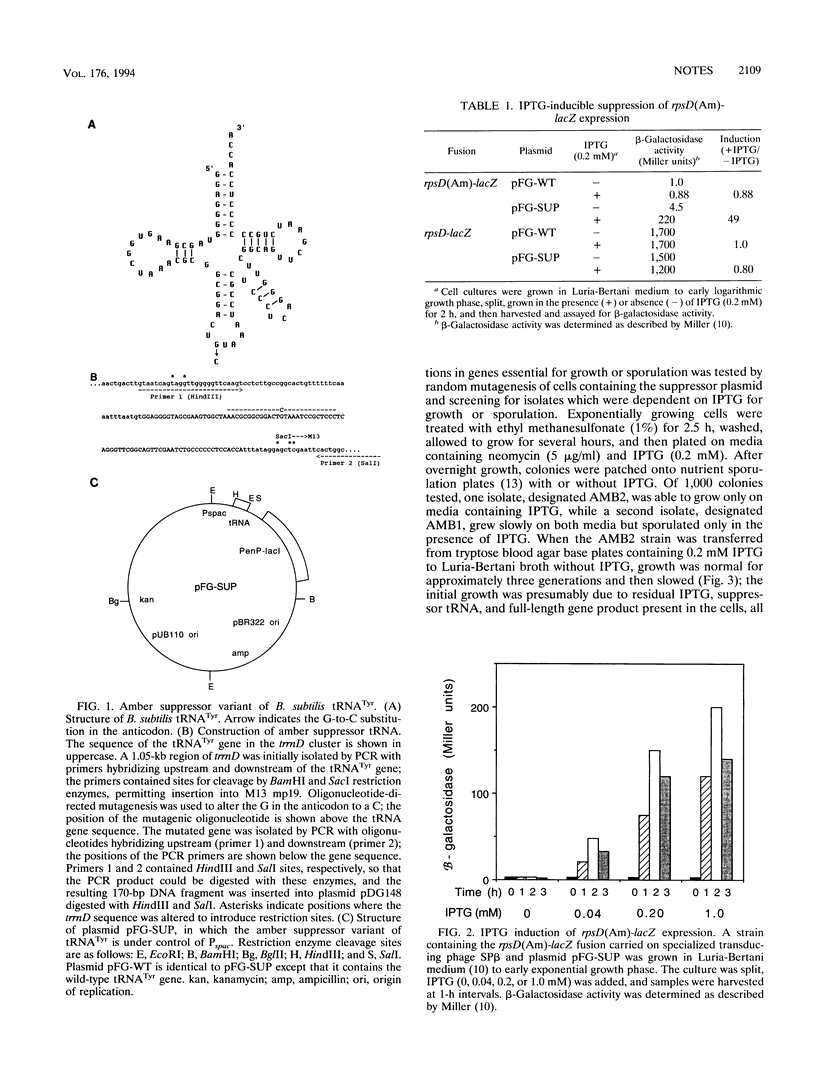
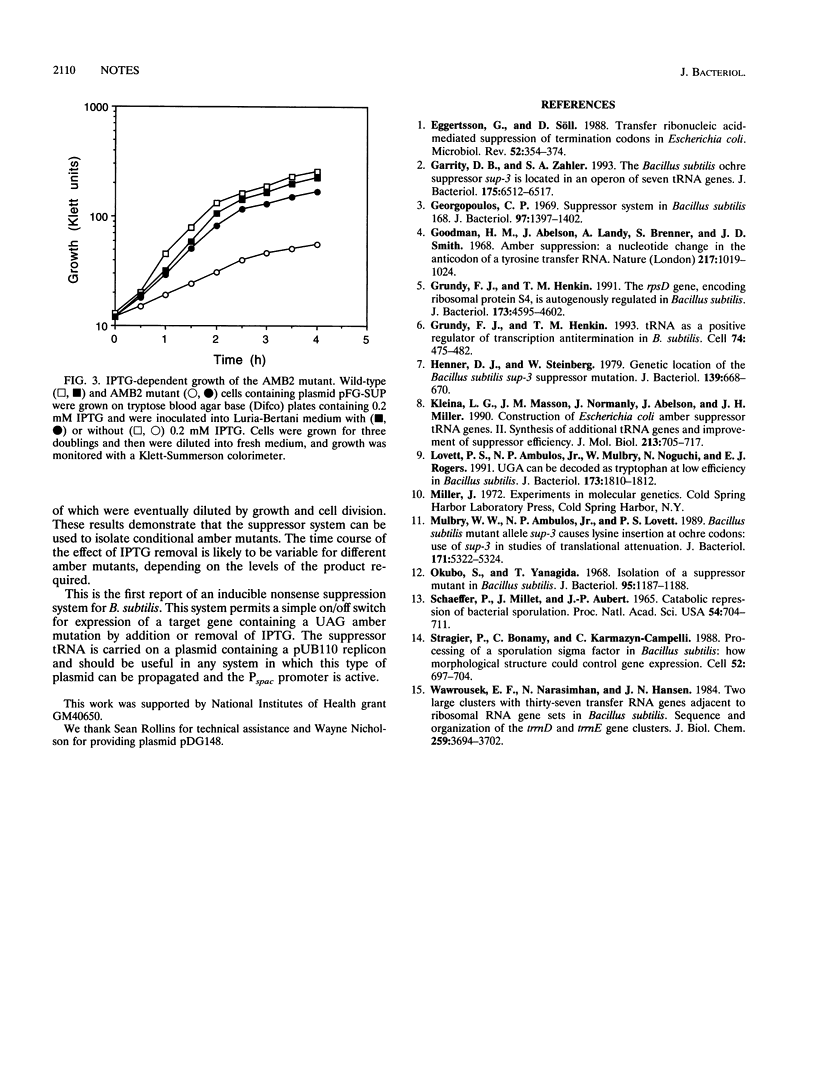
An amber suppressor variant of Bacillus subtilis tyrosyl-tRNA was constructed and placed under control of the isopropyl-beta-D-thiogalactopyranoside (IPTG)-inducible Pspac promoter. Addition of IPTG resulted in a 50-fold increase in the expression of an rpsD-lacZ fusion containing a UAG amber codon. This system permitted isolation of a conditional lethal mutant which required IPTG for growth.
Full text
PDF
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Eggertsson G., Söll D. Transfer ribonucleic acid-mediated suppression of termination codons in Escherichia coli. Microbiol Rev. 1988 Sep;52(3):354–374. doi: 10.1128/mr.52.3.354-374.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garrity D. B., Zahler S. A. The Bacillus subtilis ochre suppressor sup-3 is located in an operon of seven tRNA genes. J Bacteriol. 1993 Oct;175(20):6512–6517. doi: 10.1128/jb.175.20.6512-6517.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgopoulos C. P. Suppressor system in Bacillus subtilis 168. J Bacteriol. 1969 Mar;97(3):1397–1402. doi: 10.1128/jb.97.3.1397-1402.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman H. M., Abelson J., Landy A., Brenner S., Smith J. D. Amber suppression: a nucleotide change in the anticodon of a tyrosine transfer RNA. Nature. 1968 Mar 16;217(5133):1019–1024. doi: 10.1038/2171019a0. [DOI] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. The rpsD gene, encoding ribosomal protein S4, is autogenously regulated in Bacillus subtilis. J Bacteriol. 1991 Aug;173(15):4595–4602. doi: 10.1128/jb.173.15.4595-4602.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Henkin T. M. tRNA as a positive regulator of transcription antitermination in B. subtilis. Cell. 1993 Aug 13;74(3):475–482. doi: 10.1016/0092-8674(93)80049-k. [DOI] [PubMed] [Google Scholar]
- Henner D. J., Steinberg W. Genetic location of the Bacillus subtilis sup-3 suppressor mutation. J Bacteriol. 1979 Aug;139(2):668–670. doi: 10.1128/jb.139.2.668-670.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleina L. G., Masson J. M., Normanly J., Abelson J., Miller J. H. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J Mol Biol. 1990 Jun 20;213(4):705–717. doi: 10.1016/S0022-2836(05)80257-8. [DOI] [PubMed] [Google Scholar]
- Lovett P. S., Ambulos N. P., Jr, Mulbry W., Noguchi N., Rogers E. J. UGA can be decoded as tryptophan at low efficiency in Bacillus subtilis. J Bacteriol. 1991 Mar;173(5):1810–1812. doi: 10.1128/jb.173.5.1810-1812.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulbry W. W., Ambulos N. P., Jr, Lovett P. S. Bacillus subtilis mutant allele sup-3 causes lysine insertion at ochre codons: use of sup-3 in studies of translational attenuation. J Bacteriol. 1989 Oct;171(10):5322–5324. doi: 10.1128/jb.171.10.5322-5324.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okubo S., Yanagida T. Isolation of a suppressor mutant in Bacillus subtilis. J Bacteriol. 1968 Mar;95(3):1187–1188. doi: 10.1128/jb.95.3.1187-1188.1968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaeffer P., Millet J., Aubert J. P. Catabolic repression of bacterial sporulation. Proc Natl Acad Sci U S A. 1965 Sep;54(3):704–711. doi: 10.1073/pnas.54.3.704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stragier P., Bonamy C., Karmazyn-Campelli C. Processing of a sporulation sigma factor in Bacillus subtilis: how morphological structure could control gene expression. Cell. 1988 Mar 11;52(5):697–704. doi: 10.1016/0092-8674(88)90407-2. [DOI] [PubMed] [Google Scholar]
- Wawrousek E. F., Narasimhan N., Hansen J. N. Two large clusters with thirty-seven transfer RNA genes adjacent to ribosomal RNA gene sets in Bacillus subtilis. Sequence and organization of trrnD and trrnE gene clusters. J Biol Chem. 1984 Mar 25;259(6):3694–3702. [PubMed] [Google Scholar]


