Abstract
Bloom syndrome and Werner syndrome are genetic disorders in which an increased rate of chromosomal abnormality is observed. The genes responsible for these diseases, BLM and WRN, have been cloned and identified as homologs of the Escherichia coli recQ genes. We studied the effect of recQ mutations on illegitimate recombination, which is an aberrant biological event related to the chromosomal abnormality in humans, and found that a variety of recQ mutations increased spontaneous illegitimate recombination by 20- to 300-fold and increased UV light-induced illegitimate recombination by 10- to 100-fold. Most λbio or λpro transducing phages are formed by the recombination events at several hot spots, which are enhanced by the recQ mutation. The analysis of nucleotide sequences at the recombination junction in the transducing phages indicates that recombination at the hot spot sites as well as the non-hot spot sites takes place between short homologous sequences. Enhancement of the recombination in the recQ mutants also occurs in the recA, recBC sbcBC, or recBC sbcA backgrounds, indicating that these recombination events are mediated by none of the known recombination pathways, RecBC, RecF, and RecE. We therefore concluded that the RecQ function suppresses illegitimate recombination that depends on short homologous regions. We discuss a model, based on the 3′-to-5′ helicase activity of RecQ, to explain the role of this protein as a suppressor of illegitimate recombination.
Keywords: λ transducing phage, short homologous region, DNA rearrangement, BLM, WRN
The Escherichia coli recQ gene encodes a DNA helicase with an activity that unwinds duplex DNA in the 3′-to-5′ direction (1), and recQ is known to play an important role in homologous recombination by the RecE and RecF pathway (2, 3). A recent important finding is that mutations in the human homologs of the recQ gene, BLM and WRN, exhibit genetic disorders that are recognized as Bloom syndrome and Werner syndrome, respectively (4, 5).
Bloom syndrome is a rare autosomal-recessive disorder that exhibits short stature, neoplasia, immunodeficiency, and an increasing risk of cancer. At the cellular level, Bloom syndrome shows a striking level of genomic instability, as evidenced by an increased rate of sister chromatid exchange and chromosomal aberration (6). Werner syndrome is also a rare genetic disorder and is characterized by the premature appearance of normal aging in young adults (7). In primary fibroblast culture and in lymphocytes of patients with Werner syndrome, cells exhibit an increased rate of somatic mutations and chromosomal abnormalities such as translocations, inversions, and chromosome losses (8). Simian virus 40-transformed fibroblast cells derived from patients with Werner syndrome showed an increased rate of mutations, and the predominant form of the mutations are deletions that are shown to be generated by illegitimate recombination (9, 10). Recently, the genes responsible for the Bloom and Werner syndromes in humans have been identified as BLM and WRN, respectively. In addition to these two homologs, a third homolog, RECQL, has also been cloned (11, 12).
In this study, we have analyzed the effect of E. coli recQ mutations on illegitimate recombination to determine if the mutations produce chromosomal abnormalities similar to those in humans. Using the λ Spi− assay (13), we found that a recQ mutation enhances the frequency of illegitimate recombination and concluded that the RecQ function plays a suppressive role in illegitimate recombination in E. coli.
MATERIALS AND METHODS
Bacterial Strains, Plasmids, and Phage Strains.
All bacterial strains used are derivatives of E. coli K12; the strains are shown in Table 1. pT7Blue(R)T-Vector (Novagen) was used for cloning of Taq polymerase-amplified DNA carrying a 3′ dA overhang. pJK2041(mini-F-Kmr-recQ+) was constructed by ligating the BamHI–EcoRI recQ+ DNA fragment with the BamHI–EcoRI fragment of a mini-F vector, pJK289 (17). The BamHI–EcoRI DNA fragment containing the recQ+ gene was prepared by PCR with the oligonucleotides 5′-CCGGATCCATGAACGGTTGAGTGGTTGG-3′ and 5′-CCGAATTCACATACATTTGACTCGCGGG-3′ as primers, chromosomal DNA of an E. coli strain C600 as a template, and TaKaRa Ex Taq (Takarashuzo, Kyoto, Japan) as a polymerase.
Table 1.
E. coli strains used in this study
| Strains | Relevant genotype or phenotype | Source/construction |
|---|---|---|
| Ymel | supE supF | Our collection |
| WL95 | (P2) | Amersham |
| HI1547 | sup0 (λ†) | Ref. 13 |
| HI2033 | sup0 recQ61∷Tn3(λ†) | HI1547 × P1·KD1996(2) |
| HI2034 | sup0 recQ1802∷Tn3(λ†) | HI1547 × P1·KD2221(2) |
| HI2035 | sup0 recQ1803∷Tn3(λ†) | HI1547 × P1·BIK1224(3) |
| HI2050 | sup0 recQ1802∷Tn3ΔrecA306∷tetr(λ†) | HI2034 × P1·GY787L |
| HI1453* | recB21 recC22 sbcA23 (λ†) | JC8679(14) |
| HI1515* | recB21 recC22 sbcA23 recQ1803∷Tn3 (λ†) | BIK1224(3) |
| HI1458* | recB21 recC22 sbcB15 sbcC201 (λ†) | JC7623(15) |
| HI1531* | recB21 recC22 sbcB15 sbcC201 recQ1803∷Tn3 (λ†) | HI1458 × P1·BIK1224(3) |
| HI2055 | sup0recQ1802∷Tn3 (λ†) mini-F-Kmr | HI2034 |
| HI2056 | sup0recQ1802∷Tn3 (λ†) mini-F-Kmr-recQ+ | HI2034 |
| HI1793 | HfrH Δ(gal-attB-uvrB)proB∷λ† | HN447(16) |
| HI2002 | HfrH Δ(gal-attB-uvrB)recQ1802∷Tn3 proB∷λ† | HI1793 × P1·KD2221(2) |
*Derivatives of E. coli AB1157.
All λ prophages described here contain the cI857 mutation.
Media and Conditions of Growth of Bacteria and Phages.
Media were prepared as described in ref. 18. λYP broth was used to induce λ prophage and to prepare λ Spi− phages from single plaques. λ trypticase agar was used to titrate λ Spi− phages. λ agar was used to titrate total λ phages.
Measurement of Frequency of the λ Spi− Phages Induced Spontaneously or Induced by UV Light.
E. coli 594 λ cI857 or its derivative was grown to 2 × 108 cells per ml at 30°C. If necessary, 2 ml of the culture was irradiated for 5–20 sec (a dose of 25–100 J/m2) with a 15-W germicidal lamp. The heat induction of λ prophage was then carried out by incubation at 42°C for 15 min with aeration. The culture was then incubated at 37°C for 2 hr. The titer of λ Spi− phage was measured by plating on E. coli WL95 P2 lysogen. The titer of total phage was measured by plating on E. coli YmeI. The frequency of λ Spi− phage was obtained by dividing the titer of λ Spi− phage by the titer of total phage. To analyze recombination junctions of λ Spi− phages, independent isolation of these phages was carried out as described by Yamaguchi et al. (19).
Determination of the Structure of λ Spi− Phage by PCR.
The structure of λ Spi− phage was determined by PCR analysis of phage DNA as described by Shimizu et al. (18). First, we identified λbio or λpro transducing phages using two sets of primer oligonucleotides: #184 (base pairs 27411–27430 of λ DNA) and bio-in (base pairs 210–191 of the attB site), or #184 and #631 (base pairs 438–417 of E. coli proB). Next, the location of recombination junctions in the λbio transducing phages were determined by PCR with several primers described by Ukita and Ikeda (20). The locations of recombination junctions in the λpro transducing phages were determined by PCR with the following sets of primers: #632 (base pairs 1814–1794 of E. coli pro), #633 (base pairs 2941–2921 of E. coli pro), #634 (base pairs 4110–4090 of E. coli pro), and the primers described by Ukita and Ikeda (20). The amplified DNA fragment was cloned into the pT7 Blue(R)T vector and sequenced with a Pharmacia AFL automatic DNA sequencer.
RESULTS
The Effect of recQ Mutations on the Formation of λ Spi− Transducing Phages.
To examine the effect of recQ mutations on illegitimate recombination, we measured the frequency of formation of λbio transducing phage, which was selected indirectly by plaque formation on an E. coli P2 lysogen (Spi− phenotype) to avoid defective transducing phage (21). λ Spi− phages are rarely detected in lysates produced by thermal induction of an E. coli λ lysogen, but λ Spi− phage production is greatly enhanced if the cells are irradiated with UV light before induction (13).
E. coli HI1547 λ cI857 or HI2034 recQ1802 λ cI857 lysogens grown to mid-log phase were irradiated with several doses of UV light. The irradiated or unirradiated lysogens were heat-induced at 42°C and then incubated at 37°C. In the unirradiated condition, the frequency of formation of λ Spi− phages was found to be 22-fold higher in the recQ mutant than that in the wild type (Table 2, experiment 1). Experiments with other recQ mutants, HI2033 recQ61::Tn3 or HI2035 recQ1803::Tn3, showed that the frequency of formation of λ Spi− phages in the recQ mutant is also 48- or 36-fold higher than that in the wild type. When the bacteria were irradiated with UV light, the frequency of formation in the recQ mutant was again 10- to 20-fold higher than that in the wild type (Table 2, experiment 1).
Table 2.
Effect of a recQ mutation on spontaneous or UV light-induced fromation of λ Spi− phage
| UV dose, J/m2 | Strain | Relevant mutation | Frequency of Spi− phages (10−7) per total λ phages (SE) | Frequency relative to control | Burst size |
|---|---|---|---|---|---|
| Experiment 1 | |||||
| 0 | HI1547* | wild type | 0.053 (0.009) | 1 | 94 |
| 0 | HI2034* | recQ1802 | 0.52 (0.11) | 22 | 47 |
| 25 | HI1547* | wild type | 0.99 (0.04) | 1 | 86 |
| 25 | HI2034* | recQ1802 | 17 (2.5) | 17 | 48 |
| 50 | HI1547* | wild type | 2.2 (0.50) | 1 | 72 |
| 50 | HI2034* | recQ1802 | 21 (2.5) | 9.5 | 35 |
| Experiment 2 | |||||
| 0 | HI1793† | wild type | 0.65 (0.13) | 1 | 7.3 |
| 0 | HI2002† | recQ1802 | 190 (31) | 290 | 3.4 |
| 4 | HI1793† | wild type | 3.2 (0.61) | 1 | 9.3 |
| 4 | HI2002† | recQ1802 | 290 (16) | 91 | 5.2 |
| 8 | HI1793† | wild type | 5.7 (1.1) | 1 | 13 |
| 8 | HI2002† | recQ1802 | 330 (14) | 58 | 5.2 |
E. coli λ cI857 lysogen or its recQ derivative grown to 2 × 108/ml was irradiated with UV light at several doses. The lysogen was induced by incubation at 42°C for 15 min with shaking and then incubated at 37°C for 2 hr. The frequency of λ Spi− phages per total phages was measured as described in Materials and Methods. Numbers are averages of four determinations.
Strains carrying λ cI857 prophage at the attB site.
Strains carrying λ cI857 prophage at the secondary site in the proB gene.
Next, we measured the frequency of λpro transducing phage, which is produced in a strain lysogenic for λ cI857 at the secondary attachment site in the proB gene. E. coli HI1793 proB::λ cI857 or HI2002 recQ1802 proB::λ cI857 lysogens were irradiated with several doses of UV light, and the phage lysate was prepared. In the unirradiated condition, the frequency of formation of λ Spi− phage was again 300-fold higher in the recQ mutant than that in the wild type (Table 2, experiment 2). When the both bacteria were irradiated with UV light, the frequency of λ Spi− phage was about 100-fold higher in the recQ mutant than it was in the wild type (Table 2, experiment 2). A lower dose of UV light was used in the experiment 2 as compared with the dose in experiment 1 because HI1793 and HI2002 contain a deletion of the uvrB region. It should be noted that the effect of the recQ mutation is more pronounced in experiment 2. But this effect would not be due to the UvrB defect in the proB::λ strains, HI1793 and HI2002, because the UvrB defect did not affect the enhancement of λbio tansducing phage formation by the RecQ defect in the attB::λ strains (data not shown). These results indicate that the recQ mutation significantly increases the formation of λbio and λpro transducing phages from the UV light-irradiated and unirradiated bacteria and suggested that the recQ gene product suppresses illegitimate recombination during the formation of λ transducing phage.
To confirm that the RecQ function suppresses illegitimate recombination, a RecQ plasmid, mini-F-Kmr-recQ+, was introduced into the HI2034 recQ1802 mutant, and the frequency of λ Spi− phage formation was measured. The introduction of mini-F-Kmr-recQ reduced the enhanced illegitimate recombination in the HI2034 recQ1802 mutant under both the irradiated and the unirradiated conditions (Table 3). This result confirmed that the RecQ protein has a suppressive function in spontaneous and UV light-induced illegitimate recombination during the formation of λ transducing phage.
Table 3.
Complementation of the RecQ defect by a recQ+ plasmid
| UV dose, J/m2 | Strain | Relevant mutation or phenotype | Frequency of Spi− phages (10−7) per total λ phages (SE) | Frequency relative to control | Burst size |
|---|---|---|---|---|---|
| 0 | HI1547 | wild type | 0.014 (0.005) | 1 | 122 |
| 0 | HI2034 | recQ1802 | 0.58 (0.07) | 41 | 51 |
| 0 | HI2055 | recQ1802 mF-Kmr | 0.61 (0.12) | 44 | 52 |
| 0 | HI2056 | recQ1802 mF-Kmr-recQ+ | 0.015 (0.004) | 1.1 | 118 |
| 50 | HI1547 | wild type | 2.9 (0.5) | 1 | 80 |
| 50 | HI2034 | recQ1802 | 32 (10) | 11 | 26 |
| 50 | HI2050 | recQ1802 mF-Kmr | 23 (7) | 7.9 | 22 |
| 50 | HI2056 | recQ1802 mF-Kmr-recQ+ | 2.7 (0.6) | 0.93 | 71 |
HI1547 λ cI857 lysogen, its recQ derivative, its recQ derivative carrying mini-F-Kmr, or its recQ derivative carrying mini-F-Kmr-recQ+ was induced as described in Table 2. The frequency of λ Spi− phages per total phages was also measured as described in Table 2. Numbers are averages of four determinations.
The recQ Mutation Preferentially Affects Illegitimate Recombination at Hot Spots.
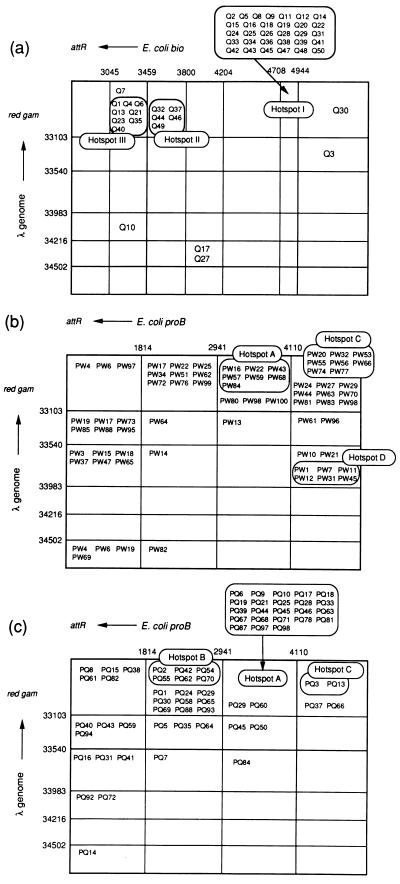
Based on the PCR analysis, we examined the distribution of recombination junctions of λ Spi− phages produced spontaneously in HI2043 recQ mutant. Many λ Spi− phages were isolated independently from the recQ mutant, and the distribution of recombination sites was estimated from locations of the junctions. Based on gel electrophoresis analysis, PCR products of the transducing phages derived from the recombination at the hot spot regions have the same sizes. Therefore, these phages appear to be formed by recombination at unique hot spots, which are confirmed by the nucleotide sequence analyses as will be described below. In the phages derived from the recQ mutant, hot spots I, II, and III, which account for 59%, 17%, and 13%, respectively, among total transducing phages tested, were found at the bio gene of E. coli and the gam-git region of λ DNA (Fig. 1a). On the other hand, in the phages derived from the wild type, the recombination at hot spot I accounts for 64% of transducing phages, but recombination events at hot spots II and III account for only 3% and <2%, respectively (data not shown). A similar result was observed when bacteria were irradiated with UV light. In the recQ mutant, the recombination events at hot spots I, II, and III account for 54%, 11%, and 18% of total transducing phages, respectively, in the irradiated condition, whereas in the wild type, recombination events at hot spots I, II, and III account for 60%, 5%, and 1% of total transducing phages, respectively, under the same condition. We therefore concluded that the recombination events at hot spots II and III are preferentially enhanced by the recQ mutation. It should be noted that the recombination event at hot spot I is also enhanced by the mutation because the frequency of total λ Spi− phage is increased 10-fold, although the percent of recombination at hot spot I in the mutant is comparable to the percent in the wild type.
Figure 1.
Distribution of recombination junctions. (a) Junctions of λbio transducing phages spontaneously induced in HI2034 recQ1802 λ lysogen. Vertical lines indicate the map coordinate of λ DNA, and horizontal lines indicate the map coordinates of E. coli bio genes. The boxes marked hot spot I, hot spot II, and hot spot III indicate groups of λbio transducing phages that are produced by the recombination at the hot spots. (b) Junctions of λpro transducing phages spontaneously induced in HI1793 wild type. Vertical lines indicate the map coordinate of λ DNA, and horizontal lines indicate the map coordinates of the proB genes. The boxes marked hot spot A, hot spot C, and hot spot D indicate groups of λpro transducing phages that are produced by the recombination at the hot spots. (c) Junctions of λpro transducing phages spontaneously induced in HI2002 recQ1802 mutant. The boxes marked hot spot A, hot spot B, and hot spot C indicate groups of λpro transducing phages that are produced by the recombination at the hot spots.
Next, we examined the distribution of recombination junctions of λ Spi− phages produced spontaneously in HI1793 proB::λ wild type and HI2002 proB::λ recQ mutant. From these strains, the transducing phage carrying the proB gene should be produced instead of λbio phage. Many λ Spi− phages were isolated independently from the wild type and recQ mutant, and the distribution of recombination sites was estimated from the locations of the junctions. In the phages derived from the wild type, many recombination events take place at hot spots A, C, or D, which were located at the proB gene of E. coli and the gam-git region of λ DNA (Fig. 1b). The recombination events at these hot spots account for 10%, 12%, and 9% of total transducing phages, respectively. On the other hand, in the phages derived from the recQ mutant, the relative frequency at hot spot A was increased 3-fold, and that at hot spot B was newly detected at the frequency of 9%, but no recombination events at hot spots C or D were detected (Fig. 1c). Therefore, the result indicates that the recombination events at hot spots A and B are relatively enhanced by the RecQ defect, but those at hot spots C and D are relatively suppressed. In conclusion, the overall frequency of illegitimate recombination was increased at hot spots I, II, III, A, and B by the RecQ defect.
A Short Region of Homology Is Required for the Recombination Enhanced by the RecQ Defect.
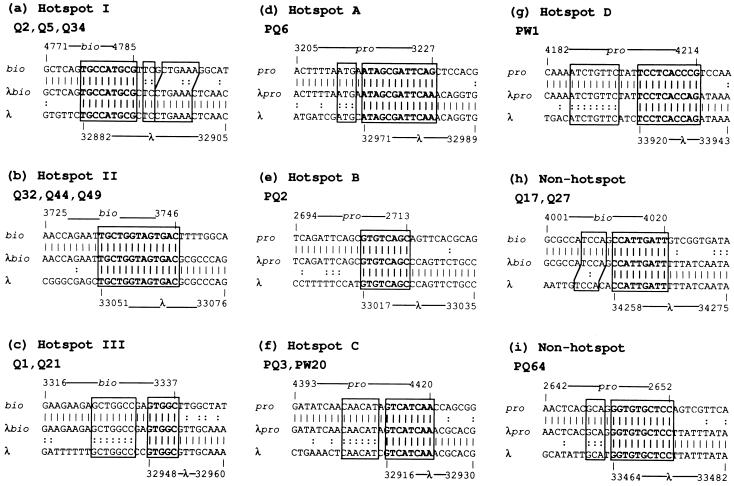
To determine junction sequences in the λbio transducing phages derived spontaneously from the recQ mutant, PCR fragments containing the junctions were cloned and sequenced. The junctions of λbio transducing phages Q2, Q5, and Q34, which were estimated to be formed by the recombination at hot spot I in Fig. 1a, were examined, and their parental recombination sites were confirmed to be the same as the hot spot found by Yamaguchi et al. (ref. 19; Fig. 2a). This hot spot was known to share a short homologous region of 9 bp in the recombination sites. Fig. 2 b and c shows the junction sequences of λbio transducing phages derived from the recQ mutant, which were formed by the recombination events at hot spots II and III. These λbio transducing phages are formed by illegitimate recombination between short regions of homology of 13 and 12 bp, respectively. Although the regions of homology at hot spot III are separated by a mismatch of 2 bp, both blocks of short regions of homology were used as crossover sites. These results indicate that short regions of homology play a role in the illegitimate recombination enhanced by the RecQ defect. It should be noted that the recombination events at the non-hot spot site also require short regions of homology. An example of those recombination events is shown in Fig. 2h. An average length of short homologous regions, including those at the hot spot and non-hot spot sites, was calculated to be 9.1 bp in the λbio transducing phages produced in the recQ mutant, a length that is comparable to those formed in the wild type (22).
Figure 2.
Nucleotide sequences of junctions of spontaneously induced λbio or λpro transducing phages. (a) Sequences of hot spot site I detected in the junctions of λbio transducing phages Q2, Q5, and Q34, which were isolated independently from unirradiated HI2034 recQ1802 λ lysogen. The sequences shown in boldface type represent homology at the recombination sites. The boxed sequences represent short regions and extra short regions of homology between the parental recombination sites. The map coordinates for phage and bacterial sequences are indicated. (b) Sequences of hot spot site II detected in the junctions of λbio transducing phages Q32, Q44, and Q49. (c) Sequences of hot spot site III detected in the junctions of λbio transducing phages Q1 and Q21. (d–g) Sequences of hot spots A, B, C, and D detected in the junctions of λpro transducing phages Q17, Q27, Q10, Q30, Q3, and Q7. (h) Sequences of a non-hot spot site detected in the junctions of λbio transducing phages Q17 and Q27. (i) Sequences of a non-hot spot site detected in the junctions of the λpro transducing phage PQ64.
Next, the junction sequences in the λpro transducing phages derived spontaneously from the wild type and recQ mutant were determined. PCR fragments containing the junctions were cloned and sequenced. Fig. 2 d–g shows the junction sequences of λpro transducing phages derived from the wild type or recQ mutant, which were formed by the recombination events at hot spots A, B, C, and D. These λpro transducing phages are formed by illegitimate recombination between short regions of homology of 12, 8, 8, and 10 bp, respectively. It is important that the recombination events at the non-hotspot site also require short regions of homology. An example of those recombination events is shown in Fig. 2i. An average length of a short homologous region, including those at the hot spot and non-hot spot sites, was calculated to be 9.0 or 8.9 bp in λpro transducing phages produced in the wild type or recQ mutant. That length is comparable to that in λbio transducing phages formed in wild type or recQ mutant. We therefore concluded that short regions of homology play a role in the illegitimate recombination enhanced by the RecQ defect.
Independence of the Enhanced Illegitimate Recombination from the Known Recombination Pathways.
To examine whether the RecA function is involved in the illegitimate recombination enhanced by the recQ mutation, the frequency of λ Spi− phages in the recQ recA double mutant was measured. The result indicates that λ Spi− phage frequency in the recQ recA double mutant was comparable to that in the recQ single mutant in both the UV light-irradiated and the unirradiated conditions (data not shown). Next, the formation of λ Spi− phages in the HI1531 recB recC sbcB sbcC recQ or the HI1515 recB recC sbcA recQ strain was compared with that of the HI1458 recB recC sbcB sbcC recQ+ strain or with that of the HI1453 recB recC sbcA recQ+ strain, respectively, in the UV light-irradiated or unirradiated condition. The recQ mutations again increased the frequency of λ Spi− phages (data not shown). These results indicate that the enhancement of illegitimate recombination by the recQ mutation takes place independently of the RecA function, the RecE pathway, or the RecF pathway.
DISCUSSION
We studied illegitimate recombination during the formation of λbio or λpro transducing phages and found that it is enhanced by the recQ mutation. Introduction of a plasmid carrying the recQ+ gene suppressed the enhanced illegitimate recombination in this mutant. These results indicate that the RecQ protein has an activity to suppress the illegitimate recombination. It has been known that the formation of λbio transducing phage is also enhanced by UV light irradiation in the wild type (13). Our result shows that the production of transducing phages is also higher in the recQ mutant than it is in the wild type in the UV light-irradiated and unirradiated conditions. Therefore, the effect of the RecQ defect and that of UV light irradiation on illegitimate recombination are synergistic.
The analyses of nucleotide sequences revealed that the frequency of illegitimate recombination at one type of hot spot is relatively enhanced by the recQ mutation and at another type of hot spot is suppressed by the mutation. Second, short regions of homology are detected in both types of hotspot sites as well as in the non-hot spot sites. Third, no specific nucleotide sequence is detected in both hot spot site types. These results show that the recombination enhanced by the RecQ defect depends on short regions of homology, similar to that in the wild-type bacteria, and that the overall frequency of illegitimate recombination detected at most hot spot and non-hot spot sites is enhanced by the recQ mutation.
In the previous study, we showed that the RecJ exonuclease preferentially stimulates UV light-induced illegitimate recombination at hot spot I and suggested that the processing of DNA ends may require at least two types of exonuclease, 5′-to-3′ and 3′-to-5′, depending on the sites of double strand breaks and short regions of homology (20). One class of recombination events may require a 5′-to-3′ exonuclease, RecJ protein, and another class of recombination events may require a 3′-to-5′ exonulcease. Such processing reactions would promote the formation of single-stranded tail, thus facilitating annealing and ligation at DNA ends to produce illegitimate recombination.
There are at least two possible mechanisms for suppression of illegitimate recombination by RecQ. First, RecQ may be able to repair spontaneous or UV light-induced DNA damage and it may then suppress the introduction of double strand break in E. coli DNA, thus preventing the formation of illegitimate recombinants via an end-joining reaction. Second, RecQ may break down a recombination intermediate, which is produced by annealing of complementary single-stranded ends. If one assume that UV light irradiation causes the double strand break of DNA via DNA damage, the fact that the UV light irradiation and the RecQ deficiency show a synergistic effect on illegitimate recombination may suggest a role for the RecQ function in the step of DNA end joining. Based on the 3′-to-5′ DNA helicase activity of RecQ, it is likely that the RecQ protein may unwind an hydrogen-bonded intermediate of the DNA end joining that is formed by annealing of DNA ends with short regions of homology, thus exhibiting the suppression of the recombination.
The recQ gene is known to be responsible for homologous recombination and double strand break repair mediated by the RecF pathway (23). The biochemical analysis has suggested that RecQ protein binds to a single-stranded region of substrate DNA and then unwinds a duplex region in the 3′-to-5′ direction with respect to the single-stranded region (1). In addition, a higher concentration of RecQ is able not only to unwind a DNA substrate with a single-stranded tail but also to unwind a blunt-ended duplex. It is thought that RecQ helicase may be involved in homologous recombination by generating a 3′ single-stranded tail, which may serve as a substrate for a recA-mediated strand exchange reaction. It has also been proposed that RecJ protein may stabilize the 3′ single-stranded tail, which is produced by RecQ-mediated unwinding, degrading the complementary 5′ tail (3). In illegitimate recombination, RecQ helicase may break down a recombinant intermediate with a 3′ single-stranded tail more efficiently and other DNA species less efficiently. It is therefore possible that a variety of substrate specificities of RecQ protein may affect the extent of suppression, depending on the hot spots.
It has been reported that the expression of the recQ gene is induced by mitomycin C and that its induction depends on RecA and LexA functions, although the binding of LexA protein to the promoter region of the recQ gene has not been demonstrated (24). The fact that the expression of the recQ gene is enhanced by DNA damage is consistent with the suppressive role of the RecQ function in illegitimate recombination in terms of maintenance of genetic information in organisms.
Recently the genes BLM and WRN, which are responsible for Bloom syndrome and Werner syndrome, respectively, have been cloned (4, 5), and a central domain of these gene products is found to be closely related to the E. coli RecQ protein. Individuals with these two genetic diseases exhibit genomic instability and predisposition to developing cancer. It is noteworthy that, in primary fibroblast culture and in lymphocytes of patients with Werner syndrome, cells show an increased rate of mutation and chromosomal aberration (8). Furthermore, simian virus 40-transformed fibroblast cells derived from patients with Werner syndrome exhibit an increased rate of mutations, and a predominant form of the mutation is that of deletions which are produced by illegitimate recombination (9, 10). Our present finding of the RecQ phenotype in E. coli is consistent with these phenotypes in the Werner mutant cells. These results suggest that the function of the recQ gene family may be conserved from bacteria to higher eukaryotes. The λ Spi− phage assay for illegitimate recombination may provide a good model system for the analysis of BLM and WRN functions.
Acknowledgments
We thank Drs. A. Nishimura, H. Nakayama, I. Kobayashi, and K. Kusano for providing bacterial strains. This work was supported by Grants-in-Aid for Scientific Research on Priority Areas to H.I. from Ministry of Education, Science, Sports and Culture of Japan.
Note Added in Proof
We measured frequency of λ Spi− phages in HI2138 recB21 recC22 sbcA23 recE159 recQ1803 or HI2137 recB21 recC22 sbcB15 sbcC201 recF143 recQ1803 λ lysogen and found that the frequency was comparable to that in HI1515 recB21 recC22 sbcA23 recE+ recQ1803 or HI1531 recB21 recC22 sbcB15 sbcC201 recF+ recQ1803 λ lysogen, respectively, in both the UV light-irradiated and the unirradiated conditions. This result confirmed that the enhanced illegitimate recombination in the recQ mutants occurs independently of the RecE and RecF functions.
References
- 1.Umezu K, Nakayama K, Nakayama H. Proc Natl Acad Sci USA. 1990;87:5363–5367. doi: 10.1073/pnas.87.14.5363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Nakayama K, Irino N, Nakayama H. Mol Gen Genet. 1985;200:266–271. doi: 10.1007/BF00425434. [DOI] [PubMed] [Google Scholar]
- 3.Kusano K, Sunohara Y, Takahashi N, Yoshikura H, Kobayashi I. Proc Natl Acad Sci USA. 1994;91:1173–1177. doi: 10.1073/pnas.91.3.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ellis N A, Groden J, Ye T-Z, Straughen J, Lennon D J, Ciocci S, Proytcheva M, German J. Cell. 1995;83:655–666. doi: 10.1016/0092-8674(95)90105-1. [DOI] [PubMed] [Google Scholar]
- 5.Yu C-E, Oshima J, Fu Y-H, Wijsman E M, Hisama F, Alisch R, Matthews S, Nakura J, Miki T, Ouais S, Martin G M, Mulligan J, Schellenberg G D. Science. 1996;272:258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 6.Langlois R G, Bigbee W L, Jensen R H, German J. Proc Natl Acad Sci USA. 1989;86:670–674. doi: 10.1073/pnas.86.2.670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cerimele D, Cottoni F, Scappaticci S, Rabbiosi G, Borroni G, Sanna E, Zei G, Fraccaro M. Hum Genet. 1982;62:25–30. doi: 10.1007/BF00295600. [DOI] [PubMed] [Google Scholar]
- 8.Scappaticci S, Cerimele D, Fraccaro M. Hum Genet. 1982;62:16–24. doi: 10.1007/BF00295599. [DOI] [PubMed] [Google Scholar]
- 9.Fukuchi K-I, Martin G M, Monnat R J. Proc Natl Acad Sci USA. 1989;86:5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Monnat R J, Hackmann A F, Chiaverotti T A. Genomics. 1992;13:777–787. doi: 10.1016/0888-7543(92)90153-j. [DOI] [PubMed] [Google Scholar]
- 11.Seki M, Miyazawa H, Tada S, Yanagisawa J, Yamaoka T, Hoshino S-i, Ozawa K, Eki T, Nogami M, Okumura K, Taguchi H, Hanaoka F, Enomoto T. Nucleic Acids Res. 1994;22:4566–4573. doi: 10.1093/nar/22.22.4566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Puranam K L, Blackshear P J. J Biol Chem. 1994;269:29838–29845. [PubMed] [Google Scholar]
- 13.Ikeda H, Shimizu H, Ukita T, Kumagai M. Adv Biophys. 1995;31:197–208. doi: 10.1016/0065-227x(95)99392-3. [DOI] [PubMed] [Google Scholar]
- 14.Gillen J R, Willis D K, Clark A J. J Bacteriol. 1981;145:521–532. doi: 10.1128/jb.145.1.521-532.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Horii Z, Clark A J. J Mol Biol. 1973;80:327–344. doi: 10.1016/0022-2836(73)90176-9. [DOI] [PubMed] [Google Scholar]
- 16.Ikeuchi T, Yura T, Yamagishi H. J Bacteriol. 1975;122:1247–1256. doi: 10.1128/jb.122.3.1247-1256.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kato J, Ikeda H. Gene. 1996;170:141–142. doi: 10.1016/0378-1119(95)00865-9. [DOI] [PubMed] [Google Scholar]
- 18.Shimizu H, Yamaguchi H, Ikeda H. Genetics. 1995;140:889–896. doi: 10.1093/genetics/140.3.889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yamaguchi H, Yamashita T, Shimizu H, Ikeda H. Mol Gen Genet. 1995;248:637–643. doi: 10.1007/BF02191702. [DOI] [PubMed] [Google Scholar]
- 20.Ukita T, Ikeda H. J Bacteriol. 1996;178:2362–2367. doi: 10.1128/jb.178.8.2362-2367.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zissler J, Signer E, Schaefer F. In: The Role of Recombination in Growth of Bacteriophage Lambda: II. Inhibition of Growth by Prophage P2. Hershey A D, editor. Plainview, NY: Cold Spring Harbor Lab. Press; 1971. pp. 469–475. [Google Scholar]
- 22.Shimizu H, Yamaguchi H, Ashizawa Y, Kohno Y, Asami M, Kato J, Ikeda H. J Mol Biol. 1997;266:297–305. doi: 10.1006/jmbi.1996.0794. [DOI] [PubMed] [Google Scholar]
- 23.Nakayama H, Nakayama K, Nakayama R, Irino N, Nakayama Y, Hanawalt P C. Mol Gen Genet. 1984;195:474–480. doi: 10.1007/BF00341449. [DOI] [PubMed] [Google Scholar]
- 24.Irino N, Nakayama K, Nakayama H. Mol Gen Genet. 1986;205:298–304. doi: 10.1007/BF00430442. [DOI] [PubMed] [Google Scholar]