Abstract
Pokeweed antiviral protein (PAP), a 29-kDa protein isolated from Phytolacca americana, inhibits translation by catalytically removing a specific adenine residue from the large rRNA of the 60S subunit of eukaryotic ribosomes. In addition to its ribosome-inactivating ability, PAP has potent antiviral activity against many plant and animal viruses, including HIV. We recently described the isolation and characterization of nontoxic PAP mutants, NT123-2, which has a point mutation (E176V) in the active site that abolishes enzymatic activity, and NT124-3, which has a nonsense mutation that results in deletion of the C-terminal 25 aa (W237Stop). In vitro translation of rabbit reticulocyte lysate ribosomes was inhibited by the C-terminal deletion mutant, but not by the active site mutant. We expressed both mutants in transgenic tobacco and showed that, unlike PAP or variant PAP, neither mutant is toxic to transgenic plants. In vivo depurination of rRNA was detected in transgenic tobacco expressing variant PAP, but not in transgenic plants expressing either the active site mutant or the C-terminal deletion mutant PAP. When extracts from transgenic plants containing the mutant PAPs were exogenously applied to tobacco leaves in the presence of potato virus X (PVX), the C-terminal deletion mutant had antiviral activity, while the active site mutant had no antiviral activity. Furthermore, transgenic plants expressing low levels of the C-terminal deletion mutant showed resistance to PVX infection, while transgenic plants expressing very high levels of the active site mutant PAP were not resistant to PVX. Our results demonstrate that an intact active site of PAP is necessary for antiviral activity, toxicity, and in vivo depurination of tobacco ribosomes. However, an intact active site is not sufficient for all these activities. An intact C terminus is also required for toxicity and depurination of tobacco ribosomes in vivo, but not for antiviral activity, suggesting that antiviral activity of PAP can be dissociated from its toxicity.
Pokeweed antiviral protein (PAP), a ribosome-inactivating protein (RIP) isolated from the leaves or seeds of Phytolacca americana, catalytically removes a specific adenine residue from a highly conserved, surface-exposed, stem-loop structure in the large rRNA of eukaryotic and prokaryotic ribosomes (1, 2). PAP displays broad-spectrum antiviral activity against plant viruses, inhibiting infection by seven different viruses, each representing a different plant virus group (3). In addition, PAP effectively inhibits infection by a number of different animal viruses, including influenza virus (4), poliovirus (5), and herpes simplex virus (6). PAP has been shown to have anti-HIV and abortifacient properties comparable to those reported for trichosanthin (7).
Single-chain (type I) RIPs, like PAP, and the A chains of two-chain (type II) RIPs, like ricin, remove an adenine base by specific cleavage of the N-glycosidic bond at A4324 in rat 28S rRNA and at homologous sites on ribosomes from other organisms. Ribosomes depurinated in this manner are unable to bind the EF-2/GTP complex, and protein synthesis is blocked at the translocation step (8, 9). Positive correlations were reported between RIP-catalyzed depurination of tobacco ribosomes and antiviral activity of exogenously applied RIPs (10) and between the extent of inhibition of virus infection by PAP and the depurination of host ribosomes (11). PAP is predominantly located in the cell wall matrix of pokeweed leaf mesophyll cells (12). Thus, it has been hypothesized that local damage caused by mechanical inoculation or by vectors that transmit viruses to plants releases PAP into the cytosol, resulting in local suicide at the site of infection (12). However, studies in animal systems have suggested that the antiviral action of PAP may not be due to its inactivation of ribosomes. PAP produced only 30% inhibition of total protein synthesis in herpes simplex virus-infected cells, whereas it inhibited virus production by >90% (6). Similarly, it has been reported that PAP inhibits HIV-1 production of p24 in both T cells and macrophages at concentrations that do not adversely affect protein synthesis (7).
We showed that transgenic plants expressing PAP are resistant to a broad spectrum of viruses (13). Transgenic plants that expressed high levels of PAP had a stunted and mottled phenotype, which was related to the level of PAP expressed (13). Recently, using random mutagenesis and selection in yeast, we isolated a number of nontoxic PAP mutants (14). To determine if enzymatic activity of PAP is required for its antiviral activity, we expressed both an active site mutant and a C-terminal deletion mutant in transgenic tobacco and examined the RNA depurination activity of the PAP mutants in vivo and the susceptibility of transgenic plants to infection by potato virus X (PVX).
MATERIALS AND METHODS
Isolation of Yeast and Tobacco Ribosomes.
PAP expression was induced in yeast (Saccharomyces cerevisiae) as previously described (14). The cells were harvested by centrifugation at 6,500 rpm in a Beckman JA21 rotor for 15 min at 4°C and washed several times. The pellets were homogenized in buffer A (200 mM Tris·HCl, pH 9.0/200 mM KCl/25 mM MgCl2/25 mM EGTA/200 mM sucrose/25 mM 2-mercaptoethanol) under liquid nitrogen and centrifuged at 11,000 × g at 4°C for 20 min. The supernatants were layered over a 10-ml cushion of 1 M sucrose/25 mM Tris·HCl, pH 7.6/25 mM KCl/5 mM MgCl2, and ribosomes were pelleted at 55,000 rpm in a Beckman 70 Ti rotor for 3.5 h at 4°C. The pellets were resuspended in 25 mM Tris·HCl, pH 7.6/25 mM KCl/5 mM MgCl2 and stored at −80°C. To isolate tobacco ribosomes, 20 g of tobacco (Nicotiana tabacum cv. Samsun) leaves were homogenized in buffer A, strained through four layers of cheesecloth, and centrifuged at 15,000 × g at 4°C for 20 min. The supernatant was layered over a 10-ml cushion of 2 M sucrose/25 mM Tris·HCl, pH 7.6/20 mM KCl/5 mM MgCl2 and centrifuged at 55,000 rpm for 3.5 h in a Beckman 70Ti rotor. Pellets were resuspended in 25 mM Tris·HCl, pH 7.6/25 mM KCl/5 mM MgCl2 and stored at −80°C.
Ribosomal RNA Depurination Assay.
Yeast ribosomes (50 μg) isolated from yeast cells transformed with the parental vector, pAC55, were incubated with 10 ng of PAP (spring leaf form; Calbiochem) at 30°C for 30 min. The reaction was stopped by the addition of SDS to 0.1%. RNA was extracted from yeast ribosomes treated with PAP or yeast ribosomes isolated from cells expressing PAP or PAP mutants, treated with 1 M aniline acetate (pH 4.5) for 30 min on ice (15) and precipitated with ethanol. RNA was electrophoresed in a 4.5% urea/polyacrylamide gel and stained with ethidium bromide. Tobacco ribosomes (50 μg) were treated with 300 ng of PAP in RIP buffer (60 mM KCl/10 mM Tris·HCl, pH 7.4/10 mM MgCl2) at 37°C for 30 min in a 100-μl final volume. An equal volume of extraction buffer (50 mM Tris·HCl, pH 8.8/240 mM NaCl/20 mM EDTA/2% SDS) was added. RNA was extracted with phenol, precipitated with ethanol, and treated with aniline. Total RNA was isolated from transgenic plants by grinding leaves in Tri Reagent (Molecular Research Center, Cincinnati), which contains phenol and guanidine thiocyanate, and treated with aniline as described above. A probe complementary to the 3′ end of rRNA was constructed by synthesizing the first strand cDNA complementary to the 3′ end of rRNA and amplifying this cDNA in a PCR as described by Taylor et al. (10). The amplified DNA fragment corresponding to the 3′ end of the rRNA was gel-purified and used as a probe to detect depurination by Northern analysis.
Construction of Plant Expression Vectors and Plant Transformation.
The 772-bp SacI/HindIII fragments from the yeast plasmid NT123-2 (encoding the active site mutant PAP) and NT124-3 (encoding the C-terminal deletion mutant PAP) were cloned into pMON8443 (13) after digestion with SacI and SmaI to remove the wild-type PAP cDNA insert and to generate NT144 and NT145, respectively. NT147 was generated by replacing the 772-bp SacI/HindIII fragment in pMON8443 with the 772-bp SacI/HindIII fragment from NT124-3.
Immunoblot Analysis of PAP from Transgenic Plants.
Six leaf disks from R1 transgenic tobacco plants were ground in 250 μl of PBS with protease inhibitors (0.1 μg/ml each of antipain, aprotinin, and leupeptin). Protein extracts were separated on a 10% SDS/PAGE and transferred to nitrocellulose membrane. The blot was probed with rabbit anti-PAP antibody, followed by horseradish peroxidase-conjugated goat anti-rabbit IgG. The blots were developed by the enhanced chemiluminescence (ECL) method (Renaissance; Dupont).
Virus Resistance Analysis.
The concentration of PAP in each transgenic plant was determined by ELISA (13). The amount of total protein in each plant extract was determined by BCA protein assay kit (Pierce). Two leaves of N. tabacum cv. Samsun were inoculated with 50 μl of 1 μg/ml PVX in the presence of protein extracts containing various concentrations of PAP, variant PAP (PAP-v), C-terminal truncated PAP, or the active site mutant PAP in PBS containing 0.05% Tween-20. Virus antigen levels in systemically infected leaves were determined by ELISA as previously described (13). PAP-expressing plants from each transgenic line and wild-type tobacco plants, were inoculated on two leaves with 1 μg/ml or 0.5 μg/ml PVX. The number of lesions on the inoculated leaves were counted, and viral antigen levels in inoculated and upper leaves were measured by ELISA (13). Student’s t test was used to determine whether the mean number of lesions on each transgenic line was significantly different compared with that of the controls. Additionally, the percentage of plants showing systemic infection in each transgenic line was compared with controls using a normal approximation to the binomial for two independent samples (z-statistic; ref. 16).
RESULTS
RNA Depurination Activity of PAP Mutants in Vivo in Yeast.
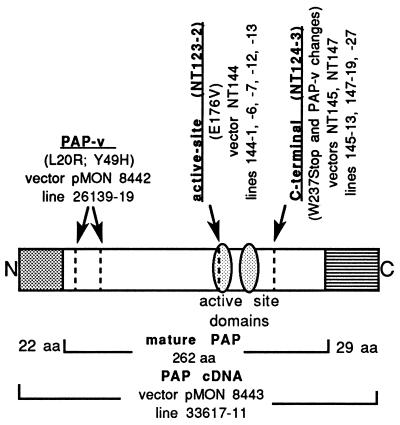
We have taken advantage of a galactose-inducible PAP expression system and the toxicity of PAP to normal yeast cells to isolate PAP mutants that are nontoxic to yeast (14). One of the mutants isolated, NT123-2, had a point mutation (E176V) at the active site, which abolished enzymatic activity in the rabbit reticulocyte lysate in vitro translation assay (14). Another mutant, NT124-3, had a nonsense mutation near the C-terminal end (W237Stop), which resulted in deletion of 25 aa from the C terminus of mature PAP. Fig. 1 shows the position of these mutations. The C-terminal deletion mutant also contains the two amino acid changes (L20R and Y49H) previously characterized in PAP-v (13).
Figure 1.
Schematic representation of the position of the mutations in PAP cDNA. NT123 is the yeast vector containing PAP cDNA and NT124 is the yeast vector containing PAP-v cDNA. The active site mutant, NT123-2, contains the point mutation E176V at the active site, the C-terminal deletion mutant, NT124-3, contains the point mutation W237Stop, and PAP-v contains two amino acid changes L20R and Y49H near the N terminus of mature PAP. Plant transformation vectors and transgenic tobacco lines are shown.
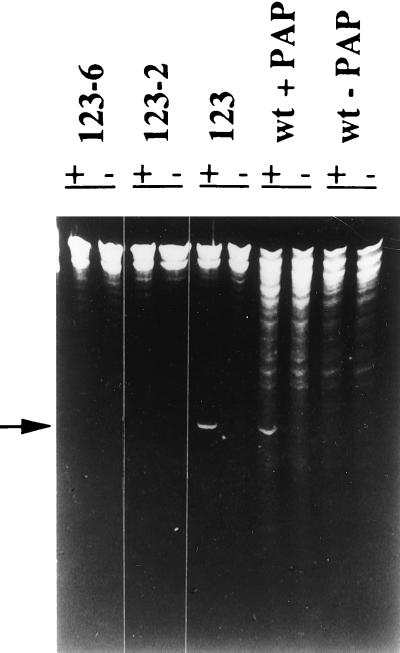
RIP-mediated depurination of the large ribosomal subunit RNA results in susceptibility of the RNA sugar–phosphate backbone to hydrolysis at the depurination site. Hence, when depurinated rRNA is treated with aniline, a small fragment is released, which is 367 nt in the yeast 26S RNA (2, 17). We used the RNA depurination assay to determine if the mutant PAPs expressed in yeast are enzymatically active in vivo. Ribosomes were isolated from yeast expressing PAP (NT123), the active site mutant PAP (NT123-2) or the C-terminal deletion mutant PAP (NT123-6), which has the same W237Stop mutation as NT124-3 (14). RNA was extracted, treated with aniline, and analyzed by gel electrophoresis as shown in Fig. 2. When ribosomes from wild-type yeast were incubated with PAP and the rRNA extracted from these ribosomes was treated with aniline, an RNA fragment of ≈367 nt was released. Aniline treatment of yeast ribosomes in the absence of PAP failed to generate this fragment, indicating that yeast does not have endogenous RIP activity. This diagnostic RNA fragment was generated when rRNA isolated from cells expressing PAP (NT123) was treated with aniline, but not from cells expressing the active site mutant (NT123-2) or the C-terminal deletion mutant (NT123-6). These results indicate that the yeast ribosomes are depurinated in vivo by PAP, but not by either mutant PAP, even though the mutant PAPs are expressed at higher levels than the wild-type PAP (14).
Figure 2.
Depurination of yeast ribosomes in vivo. Ribosomes were isolated from wild-type yeast (wt), yeast expressing PAP (123), the active site mutant (123-2), and the C-terminal deletion mutant (123-6), and rRNA was extracted, treated with aniline, separated on a 4.5% urea/polyacrylamide gel, and stained with ethidium bromide. WT + PAP represents ribosomes isolated from wild-type yeast treated with 10 ng of PAP, + and − denote the presence and absence of aniline treatment, respectively, and the arrow shows the position of the diagnostic cleavage product of rRNA.
Expression of PAP Mutants in Transgenic Tobacco.
To determine if depurination of host ribosomes is required for antiviral activity, the active site mutant and the C-terminal deletion mutant PAPs were transformed into tobacco. Transformation frequencies of N. tabacum cv. Samsun typically range between 10 to 12%, based on the number of transgenic plants obtained per leaf disk (13). The transformation frequency was 13% using NT144, containing the active site mutant PAP and 11%, using NT145, containing the C-terminal deletion mutant PAP. Furthermore, transgenic plants expressing the active site mutant or the C-terminal deletion mutant (Fig. 3) were phenotypically normal. They grew at the same rate as wild-type plants and did not show chlorosis or mottling on their leaves, indicating that the expression of the mutant PAPs was not toxic to transgenic tobacco. These results are in sharp contrast to the previously reported results, in which the transformation frequencies of N. tabacum were reduced to 0.7% when a vector containing PAP (pMON8443) was used and to 3.7% when a vector containing PAP-v (pMON8442) was used, both of which are enzymatically active (13). In previous experiments, we did not recover any transgenic plants expressing high levels of PAP, and as shown in Fig. 3, plants expressing high levels of PAP-v showed growth reduction and lesions on their leaves (13).
Figure 3.
Comparison of transgenic plants expressing PAP and PAP mutants. Transgenic plants expressing PAP-v (middle) showed chlorotic lesions on their leaves, while plants expressing the C-terminal deletion mutant (right) were indistinguishable from the wild-type tobacco plants (left).
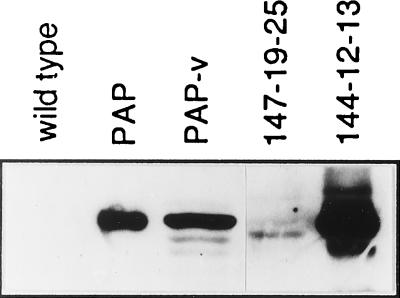
Regenerated transgenic plants were first analyzed for expression of neomycin phosphotransferase by ELISA. The neomycin phosphotransferase-positive plants were then analyzed for PAP expression by ELISA and immunoblot analysis. Eleven different transgenic plants expressed detectable levels of the active site mutant by ELISA. As shown in Fig. 4, plants expressing the active site mutant PAP produced a 29-kDa protein that comigrated with mature PAP, indicating that the active site mutant PAP is fully processed to the mature form in transgenic plants. Active site mutant PAP was expressed at significantly higher levels in transgenic plants than PAP-v. No bands corresponding to PAP were detected in wild-type tobacco (Fig. 4). A single protein of ≈27 kDa crossreacted with PAP antibodies in plants expressing the C-terminal deletion mutant. The expression levels of the C-terminal deletion mutant were variable among individual plants and significantly lower than the active site mutant (Fig. 4).
Figure 4.
Immunoblot analysis of PAP expressed in transgenic plants. Total protein was separated on a 10% SDS/polyacrylamide gel and transferred to nitrocellulose. The blot was probed with polyclonal antibody against PAP followed by anti-rabbit IgG. Lanes: wild type, 50 μg of protein from wild-type tobacco; PAP, 20 ng of purified PAP (Calbiochem); PAP-v, 50 μg of protein from transgenic line 26139-19; 147-19-25, 70 μg of protein from transgenic line 147-19-25 (C-terminal deletion mutant); and 144-12-13, 25 μg of protein from transgenic line 144-12-13 (active site mutant).
RNA Depurination Activity of PAP Mutants in Vivo in Transgenic Tobacco.
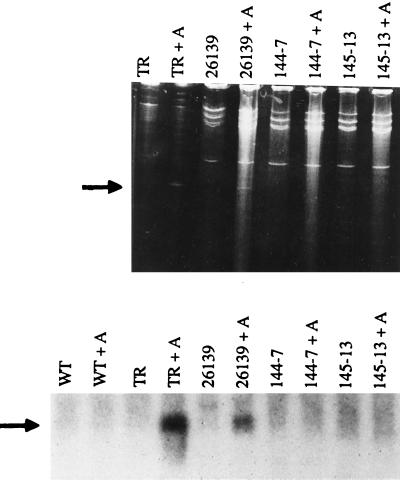
The lack of toxicity observed with the active site mutant and the C-terminal deletion mutant in transgenic tobacco suggested that neither mutant depurinated tobacco ribosomes in vivo. To determine if in vivo depurination of the rRNA can be detected in transgenic plants, total RNA was extracted from transgenic plants expressing PAP-v or PAP mutants, treated with aniline, and analyzed by electrophoresis before and after aniline treatment, as shown in Fig. 5 Upper. When ribosomes isolated from wild-type tobacco plants were treated with 90 nM PAP and rRNA extracted from these ribosomes was treated with aniline, an RNA fragment of ≈360 nt was released. It has been shown that a similar fragment is generated by depurination at the adenine residue corresponding to A4324 in rat 28S rRNA (18). PAP treatment of tobacco ribosomes in the absence of aniline failed to generate this fragment. The 360-nt RNA fragment was generated from aniline-treated total RNA isolated from the leaves of transgenic tobacco plants expressing PAP-v, but not from transgenic plants expressing either the active site mutant or the C-terminal deletion mutant. Intact rRNA can be isolated from pokeweed using this method, indicating that depurination does not occur during the RNA isolation protocol (data not shown).
Figure 5.
Depurination of tobacco ribosomes in vivo. Ribosomes were isolated from wild-type tobacco plants and 50 μg of ribosomes were treated with 300 ng of PAP as described. RNA was extracted (TR) and divided in half, and one half was treated with aniline (TR + A). Total RNA (20 μg) isolated from transgenic plants expressing PAP-v (26139), the active site mutant (144-7), the C-terminal deletion mutant (145-13), and wild-type tobacco plants (WT) was divided in half, and one half was treated with aniline (+ A). RNAs were separated on a 4.5% urea/polyacrylamide gel and visualized after staining with ethidium bromide (Upper) or were separated on a 1.3% formaldehyde agarose gel and probed with a PCR fragment complementary to the 3′ end of the rRNA (Lower). The position of the 360-nt diagnostic cleavage product of the rRNA is indicated with an arrow.
To determine if depurination occurs at levels that cannot be detected by urea/acrylamide gel analysis, we used Northern blot analysis to examine depurination. RNA samples were probed with a PCR fragment corresponding to the 3′ end of tobacco 28S rRNA. As shown in Fig. 5 Lower, aniline treatment of RNA extracted from PAP-treated tobacco ribosomes produced the diagnostic RNA fragment that hybridized to the probe. The diagnostic fragment was observed with aniline-treated total RNA from transgenic plants expressing PAP-v, but not with aniline-treated RNA from wild-type tobacco plants or from plants expressing either mutant PAP. The concentration of PAP that gives 50% depurination of tobacco ribosomes in the in vitro depurination assay is 4 ng/ml (10). The expression levels of PAP-v in line 26139-19 (10 ng/ml), the C-terminal deletion mutant PAP in line 145-13 (5 ng/ml), and the active site mutant PAP in line 144-7 (50 ng/ml) are within the limits of the depurination assay. These results demonstrate that ribosomes are depurinated in vivo in transgenic plants expressing PAP-v, but not in plants expressing either mutant PAP within the limits of the assay.
Antiviral Activity of Exogenously Applied Mutant PAPs.
To test if mutant PAPs have antiviral activity, wild-type tobacco plants were inoculated with PVX in the presence of protein extracts from transgenic plants expressing the mutant PAPs. Tobacco plants inoculated with 1 μg/ml PVX in the presence of different amounts of total protein (ranging from 6.7 μg to 1.1 mg) from nontransformed plants became infected with PVX and showed local lesions, indicating that protein extracts from nontransformed tobacco have no inhibitory effect on PVX infection. To demonstrate the effect of PAP on PVX infection, protein extracts from nontransformed tobacco plants were applied to tobacco leaves in the presence of 5 and 10 ng of purified PAP. This resulted in a dramatic reduction in the numbers of PVX lesions on inoculated leaves compared with the control (Table 1). Tobacco plants were also inoculated with PVX in the presence of protein extracts from transgenic plants expressing PAP-v, the active site mutant, and the C-terminal deletion mutant PAP. PAP levels in the extracts were quantitated by ELISA, and the amount was adjusted so that known amounts of PAP could be applied to test plants. A dramatic reduction in the number of PVX lesions was observed on inoculated leaves treated with 5 ng of PAP-v. Similarly, when PVX was inoculated in the presence of 5 ng of C-terminal deletion mutant PAP from transgenic plant 145-13, a reduction in the numbers of PVX lesions was observed. In contrast, when PVX was inoculated in the presence of 5–100 ng of active site mutant PAP from transgenic plant 144-12, the numbers of lesions observed on the inoculated leaves were similar to the control plants. These results demonstrate that exogenous application of the active site mutant PAP does not protect tobacco from PVX infection, while exogenous application of PAP-v or the C-terminal deletion mutant PAP protects tobacco from PVX.
Table 1.
Susceptibility of tobacco plants to PVX in the presence of exogenously applied PAPs
| Plant extract* | PAP expressed | PAP applied,† ng per leaf | Mean no. of lesions‡ |
|---|---|---|---|
| WT | 0 | 66.6 ± 10.1 | |
| WT plus PAP§ | 5 | 9.0 ± 2.0 | |
| 10 | 1.5 ± 2.0 | ||
| 26139-19 | PAP-v | 5 | 1.8 ± 2.9 |
| 145-13 | W237Stop | 5 | 12.5 ± 7.4 |
| 144-12 | E176V | 5 | 57.8 ± 7.4 |
| 10 | 56.1 ± 4.9 | ||
| 20 | 55.5 ± 13.8 | ||
| 50 | 53.3 ± 14.9 | ||
| 100 | 68.0 ± 11.7 |
WT, wild type.
Protein extracts were prepared from either nontransformed (WT) or transformed tobacco leaves expressing PAP-v (26139-19), the active site mutant (144-12), or the C-terminal deletion mutant (145-13). Twenty plants for WT, ten plants for WT plus PAP, and five plants for each transgenic protein extract were used in the assay.
PAP levels were quantitated by ELISA and indicated amounts of PAP was applied to two leaves from each plant, together with 1 μg/ml PVX.
The number of lesions were counted at 9 days postinoculation. Mean values ± SE for the total number of lesions per plant are shown.
Purified PAP (Calbiochem) was added to a protein extract from nontransformed tobacco leaves.
Antiviral Activity of the Active-Site Mutant in Transgenic Tobacco.
To determine if transgenic lines expressing the active site mutant PAP are resistant to virus infection, self-fertilized R1 progeny from four different independently transformed N. tabacum lines were screened for the presence of PAP by ELISA, and PAP-positive plants were used in virus resistance tests. Ten progeny from each transgenic line expressing the active site mutant PAP, PAP-v, and PAP and 10 nontransformed tobacco plants were inoculated with 1 μg/ml PVX. Symptom development on both inoculated and upper leaves was monitored visually each day up to 21 days postinoculation. In addition, the inoculated and the first, second, and third systemically infected leaves of each plant were sampled at 12 days postinoculation to quantitate virus replication and spread. As shown in Table 2 (top rows), transgenic plants expressing PAP-v or PAP did not develop any lesions on their inoculated leaves at 9 days postinoculation. In contrast, transgenic plants expressing the active site mutant had as many lesions on their inoculated leaves as the control plants. ELISA analysis showed that 90% of wild-type tobacco plants were systemically infected by PVX at 12 days postinoculation, while only 30 and 40% of the transgenic plants expressing PAP-v and PAP, respectively, were infected. In contrast, 100% of the plants expressing the active site mutant were infected with PVX (Table 2).
Table 2.
Susceptibility of transgenic tobacco plants expressing the active site mutant PAP (E176V) to PVX infection
| Plant line | PAP expressed | PAP* ng/mg | No. of lesions† | % infected‡ |
|---|---|---|---|---|
| WT | 0 | 77 ± 4 | 90 | |
| 26139-19 | PAP-v | 5.6 ± 2.6 | 0§ | 30§ |
| 33617-11 | PAP | 0.6 ± 0.02 | 0§ | 40¶ |
| 144-1 | E176V | 43.8 ± 4.8 | 78 ± 5 | 100 |
| 144-7 | E176V | 46.2 ± 5.6 | 72 ± 4 | 100 |
| WT | 0 | 12 ± 2 | 100 | |
| 26139-19 | PAP-v | 9.6 ± 2.6 | 1 ± 0.3§ | 30§ |
| 144-6 | E176V | 1,045 ± 45 | 11 ± 2 | 100 |
| 144-13 | E176V | 900 ± 50 | 13 ± 3 | 100 |
Twenty plants from R1 progeny of each transgenic line were analyzed ELISA for expression of PAP and PAP-positive plants were used in virus resistance tests. Mean values ± SE are shown for each transgenic line.
Ten plants from each line were inoculated with 1 μg/ml PVX (top rows) or 0.5 μg/ml PVX (bottom rows) and the number of lesions were counted 9 days postinoculation (dpi). Mean values ± SE of the mean for the numbers of lesions per plant are shown.
Three leaf disks were taken from first, second, and third systemically infected leaves at 12 dpi, and viral antigen levels were quantitated by ELISA.
Significantly different from wild type at 1% level.
Significantly different from wild type at 5% level.
To determine if transgenic plants expressing higher levels of the active site mutant are also susceptible to PVX infection, R1 progeny of two transgenic lines expressing higher levels of PAP were inoculated with 0.5 μg/ml PVX (Table 2, bottom rows). Both of these lines had similar numbers of lesions on their inoculated leaves as the wild-type tobacco plants and similar percentages of these plants showed systemic infection as the wild-type controls. To determine if the lack of resistance observed in the R1 progeny of a total of seven different transgenic lines tested, is observed in the homozygous progeny, we analyzed R2 progeny transgenic line 144-12, which expressed the highest levels of the active site mutant. As shown in Table 3 (bottom rows), transgenic plants producing high levels of the active site mutant had the same numbers of lesions as the wild-type tobacco plants in their inoculated leaves, and by 12 days postinoculation, 100% of the transgenic plants expressing the active site mutant were systemically infected with PVX.
Table 3.
Susceptibility of transgenic tobacco plants expressing the C-terminal deletion mutant (W237Stop) to PVX infection
| Plant line | PAP expressed | Level of PAP,* ng/mg | No. of lesions† | % of plants showing systemic infection‡
|
|
|---|---|---|---|---|---|
| 12 dpi | 21 dpi | ||||
| WT | 0 | 62 ± 10 | 80 | 80 | |
| 26139-19 | PAP-v | 9.6 | 21 ± 8§ | 30¶ | 30¶ |
| 147-27 | W237Stop | 2.0 | 24 ± 6§ | 30¶ | 30¶ |
| 147-19 | W237Stop | 3.8 | 10 ± 3§ | 70 | 70 |
| WT | 0 | 24 ± 5 | 90 | 90 | |
| 26139-19 | PAP-v | 9.6 | 1 ± 0.6§ | 10§ | 30§ |
| 33617-11 | PAP | 1.6 | 11 ± 1§ | 10§ | 40¶ |
| 144-12-13 | E176V | 1,500 | 23 ± 4 | 100 | 100 |
| 147-19-25 | W237Stop | 3.8 | 12 ± 3¶ | 20§ | 60 |
| 145-13-19 | W237Stop | 4.4 | 6 ± 1§ | 30§ | 60 |
dpi, Days postinoculation.
PAP levels were quantitated by ELISA in the primary transgenic plants. Only PAP-positive plants were used in virus resistance tests.
Ten plants from progeny (top rows, R1 generation; bottom rows, R2 generation) of each transgenic line were inoculated with 50 μl of 1 μg/ml PVX (top rows) or 0.5 μg/ml PVX (bottom rows) on two leaves per plant. The number of lesions were counted 12 dpi. Mean values ± SE of the mean are shown for the number of lesions per plant. The results are from two representative experiments that were repeated three times with similar results.
Two leaf disks were taken from first and second systemically infected leaf from each plant at 12 dpi, and two leaf disks were taken from third and fourth systemically infected leaf at 21 dpi. Viral antigen levels were quantitated by ELISA.
Significantly different from wild type at 1% level.
Significantly different from wild type at 5% level.
Antiviral Activity of C-Terminal Deletion Mutant in Transgenic Tobacco.
To determine if transgenic lines expressing the C-terminal deletion mutant are resistant to virus infection, self-fertilized progeny (R1 generation) from two different transgenic lines expressing the C-terminal deletion mutant PAP were inoculated with 1 μg/ml PVX. As shown in Table 3 (top rows), both lines had significantly lower numbers of lesions on their inoculated leaves compared with the wild-type plants. By 21 days postinoculation, only 30% of the plants from the progeny of lines 26139-19 and 147-27 contained PVX antigen by ELISA, while 70% of the plants from line 147-19 were infected with PVX.
To determine if homozygous progeny expressing the C-terminal deletion mutant are resistant to PVX, homozygous progeny (R2 generation) from lines 147-19 and 145-13 were inoculated with 0.5 μg/ml PVX, and the numbers of lesions were counted at 12 days postinoculation. As shown in Table 3 (bottom rows), plants from transgenic lines 147-19-25 and 145-13-19 had significantly lower numbers of lesions on their inoculated leaves compared with the wild-type plants, and at 12 days postinoculation, only 20–30% of the these plants showed systemic symptoms and contained PVX antigen by ELISA, while 90% of the control plants were infected with PVX. At 21 days postinoculation, 60% of plants from lines 147-19-25 and 145-13-19 were infected; however, PVX antigen levels were 3 and 2.5 times lower in plants from lines 147-19-25 and 145-13-19, respectively, compared with the antigen levels in wild-type plants. A total of six different independently transformed transgenic lines expressing the C-terminal deletion mutant was analyzed. Two lines showed reduced incidence of infection, two lines showed a delay against PVX infection, and two lines were susceptible to PVX.
DISCUSSION
To understand the basis for cytotoxicity and antiviral activity of PAP, we expressed an active site mutant and a C-terminal deletion mutant in transgenic tobacco. Transgenic plants expressing either mutant were indistinguishable from wild-type tobacco plants. Depurination of rRNA was detected in transgenic plants expressing PAP-v but not in transgenic plants expressing either mutant PAP. Immunoblot analysis showed that the active site mutant PAP produced in transgenic tobacco comigrated with the mature form, indicating that the mutation did not affect N-terminal or C-terminal processing of the protein. The active site mutant did not protect tobacco plants from PVX infection even when it was exogenously applied to tobacco leaves at 20-fold higher concentrations than PAP-v. Furthermore, seven different independently transformed lines expressing the active site mutant PAP did not show resistance to PVX infection. Since several different independently transformed lines were susceptible to PVX, the lack of resistance observed cannot be due to somaclonal variation during tissue culture. These results conclusively demonstrate that an intact active site of PAP is needed for antiviral activity, toxicity, and in vivo depurination of tobacco ribosomes. This is the first demonstration that an intact active site is required for the antiviral activity of a plant RIP. Our results also show that an intact active site is not necessarily sufficient for all these activities. Transgenic plants expressing the C-terminal deletion mutant PAP were phenotypically normal, and depurination of rRNA was not detected in these plants. In contrast to the active site mutant, the C-terminal deletion mutant was antiviral when exogenously applied, and several transgenic lines expressing this mutant showed enhanced resistance to viral infection. These results demonstrate that the C-terminal sequences of PAP are required for toxicity and in vivo depurination of tobacco ribosomes, but not for antiviral activity.
The three-dimensional organization of the putative active site region is highly conserved between PAP and the ricin A chain (19). However, ricin and PAP differ significantly in their substrate specificity. PAP can depurinate ribosomes from all eukaryotes and prokaryotes, while only mammalian ribosomes are extremely sensitive to depurination by the ricin A chain (20). By making hybrids between different domains of PAP and the ricin A chain, Chaddock et al. (21) recently showed that sequences outside the active site of PAP are involved in recognition of prokaryotic ribosomes. Studies with ricin have also shown that an intact active site is not itself sufficient for antiviral activity, since ricin A chain is not antiviral even though PAP and ricin have similar residues at their active site (10). Recently, saporin-L1, a single chain RIP from the leaves of Saponaria officinalis, has been reported to release adenine residues from various RNAs and DNA, including tobacco mosaic virus genomic RNA (22). PAP has a putative RNA-binding domain near its N terminus (residues 70–76; ref. 23) and can recognize PVX RNA as a substrate (unpublished data). If antiviral activity of PAP is due to depurination of the viral RNA, then residues involved in RNA recognition and catalytic activity would be critical. It is unlikely that the E176V mutation in PAP would affect both substrate binding and catalytic activity, since mutation of the corresponding Glu in ricin (E177A) affects the catalytic rate (kcat) but not the substrate binding (Km), suggesting that Glu-177 is not involved in substrate binding, but it controls the rate-limiting step in the depurination reaction (24).
Modeling the E176V mutation into the three-dimensional structure of PAP predicts that the mutant protein would fold as the wild-type protein, and, based on the proposed catalytic mechanism for PAP, E176V mutant would be predicted to be catalytically inactive (A. Monzingo and J. Robertus, personal communication). Analysis of the subcellular localization of PAP and the active site mutant PAP expressed in yeast indicates that both are localized in the endoplasmic reticulum (unpublished data). Immunoblot analysis, immunolocalization studies, and molecular modeling indicate that the E176V mutation does not affect processing, stability, subcellular localization, and possibly the folding properties of PAP. These results suggest that it is unlikely that the E176V mutation in PAP affects more than one function.
The C-terminal deletion mutant was expressed at significantly lower levels than the active site mutant in transgenic plants. This mutant inhibited in vitro translation of animal ribosomes (14), yet it had no detectable ribosome specific depurination activity in vivo in yeast and in tobacco. The C-terminal deletion mutant purified from Escherichia coli depurinated tobacco ribosomes in vitro, while the active site mutant purified from E. coli did not (unpublished data). Possible explanations for the reduction or lack of antiribosomal activity of the C-terminal deletion mutant in vivo include a change in subcellular localization, which reduces contact between ribosomes and the mutant PAP, a change in substrate affinity or protein stability. Indirect immunofluorescence studies in yeast indicate that subcellular localization of the C-terminal deletion mutant might be altered. Modeling the W237Stop mutation into the three-dimensional structure of PAP using the program x-plor, predicts that this mutant would be ≈10% less stable than the wild-type PAP (A. Monzingo and J. Robertus, personal communication). Although the C-terminal deletion mutant might have undetectable antiribosomal activity in vivo, immunolocalization studies and molecular modeling suggest that deletion of the C-terminal 25 aa might affect the subcellular localization and the stability of the mutant PAP such that it cannot depurinate host ribosomes in vivo. Our results show that an intact C terminus is not required for antiviral activity, suggesting that the C terminus of PAP may not contain crucial viral RNA recognition determinants. Separation of antiviral activity from toxicity is significant, because it would likely provide a RIP with enhanced applications in agriculture and medicine.
Acknowledgments
We thank Drs. Peter Day, Eric Lam, and Annette Chiang for critical reading of the manuscript, Oleg Zoubenko for tobacco transformation, Chris Coetzer for constructing NT147, Li Wang for help with ELISA analysis, and Larry Holden and Anthony Carella of Monsanto Company and Chris Coetzer for statistical analysis of the data. This work was supported by National Science Foundation Grant MCB-9419919 and a Johnson & Johnson Discovery Grant to N.E.T.
ABBREVIATIONS
- PAP
pokeweed antiviral protein
- RIP
ribosome-inactivating protein
- PVX
potato virus X
- PAP-v
variant PAP
References
- 1.Endo Y, Tsurugi K, Lambert J M. Biochem Biophys Res Commun. 1988;150:1032–1036. doi: 10.1016/0006-291x(88)90733-4. [DOI] [PubMed] [Google Scholar]
- 2.Hartley M R, Legname G, Osborn R, Chen Z, Lord J M. FEBS Lett. 1991;290:65–68. doi: 10.1016/0014-5793(91)81227-y. [DOI] [PubMed] [Google Scholar]
- 3.Chen Z C, Antoniw J F, White R F, Lin Q. Plant Pathol. 1992;40:612–620. [Google Scholar]
- 4.Tomlinson J A, Walker V M, Flewett T H, Barclay G R. J Gen Virol. 1974;22:225–232. doi: 10.1099/0022-1317-22-2-225. [DOI] [PubMed] [Google Scholar]
- 5.Ussery M A, Irvin J D, Hardesty B. Ann NY Acad Sci. 1977;284:431–440. doi: 10.1111/j.1749-6632.1977.tb21979.x. [DOI] [PubMed] [Google Scholar]
- 6.Aron G M, Irvin J D. Antimicrob Agents Chemother. 1980;17:1032–1033. doi: 10.1128/aac.17.6.1032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zarling J M, Moran P A, Haffar O, Sias J, Richman D D, Spina C A, Myers D E, Kuelbeck V, Ledbetter J A, Uckun F M. Nature (London) 1990;347:92–95. doi: 10.1038/347092a0. [DOI] [PubMed] [Google Scholar]
- 8.Montanaro L, Sperti S, Mattioli A, Testoni G, Stirpe F. Biochem J. 1975;146:127–131. doi: 10.1042/bj1460127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborn R W, Hartley M R. Eur J Biochem. 1990;193:401–407. doi: 10.1111/j.1432-1033.1990.tb19353.x. [DOI] [PubMed] [Google Scholar]
- 10.Taylor S, Massiah A, Lomonossoff G, Roberts L, Lord J M, Hartley M. Plant J. 1994;5:827–835. doi: 10.1046/j.1365-313x.1994.5060827.x. [DOI] [PubMed] [Google Scholar]
- 11.Chen Z C, Antoniw J F, White R F. Physiol Mol Plant Pathol. 1993;42:249–258. [Google Scholar]
- 12.Ready M P, Brown D T, Robertus J D. Proc Natl Acad Sci USA. 1986;83:5053–5056. doi: 10.1073/pnas.83.14.5053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lodge J K, Kaniewski W, Tumer N E. Proc Natl Acad Sci USA. 1993;90:7089–7093. doi: 10.1073/pnas.90.15.7089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hur Y, Hwang D J, Zoubenko O, Coetzer C, Uckun F, Tumer N E. Proc Natl Acad Sci USA. 1995;92:8448–8452. doi: 10.1073/pnas.92.18.8448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bass H, Webster C, O’Brian G R, Roberts J K M, Boston R S. Plant Cell. 1992;4:225–234. doi: 10.1105/tpc.4.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Snedecor G W, Cochran W G. Statistical Methods. 7th Ed. Ames: Iowa State Univ. Press; 1980. pp. 201–202. [Google Scholar]
- 17.Stirpe F, Bailey S, Miller S P, Bodley J W. Nucleic Acids Res. 1988;16:1349–1357. doi: 10.1093/nar/16.4.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Prestle J, Schonfelder M, Adam G, Mundry K W. Nucleic Acids Res. 1992;20:3179–3182. doi: 10.1093/nar/20.12.3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Monzingo A F, Collins E J, Ernst S R, Irvin J D, Robertus J D. J Mol Biol. 1993;233:705–715. doi: 10.1006/jmbi.1993.1547. [DOI] [PubMed] [Google Scholar]
- 20.Irvin J D. In: Antiviral Proteins in Higher Plants. Chessin M, DeBorde D, Zipf A, editors. Boca Raton, FL: CRC; 1995. pp. 65–94. [Google Scholar]
- 21.Chaddock J A, Monzingo A F, Robertus J D, Lord J M, Roberts L M. Eur J Biochem. 1996;235:159–166. doi: 10.1111/j.1432-1033.1996.00159.x. [DOI] [PubMed] [Google Scholar]
- 22.Barbieri L, Gorini P, Valbonesi P, Castiglioni P, Stirpe F. Nature (London) 1994;372:624. doi: 10.1038/372624a0. [DOI] [PubMed] [Google Scholar]
- 23.Bass H W, Webster C, O’Brian G R, Roberts J K M, Boston R S. Plant Cell. 1992;4:225–234. doi: 10.1105/tpc.4.2.225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y, Mlsna D, Monzingo A F, Ready M P, Frankel A, Robertus J. Biochemistry. 1992;31:3294–3296. doi: 10.1021/bi00127a035. [DOI] [PubMed] [Google Scholar]