Abstract
Huntington disease (HD) is associated with abnormal expansions of a CAG repeat close to the 5′ end of the IT15 gene. We have assembled a set of 293 HD subjects whose ages of onset were known and sized their HD CAG repeats. These repeats accounted for 69% of the variance of age of onset when we used the most parsimonious model, which relates the logarithm of age of onset to a function of CAG repeat number. Since other familial factors have been proposed to influence the age of onset of HD, we have examined a number of candidate loci. The CAG repeat number on normal chromosomes, the Δ2642 polymorphism in the HD gene, and apolipoprotein E genotypes did not affect the age of onset of HD. Although mitochondrial energy production defects in HD have led to suggestions that variants in the mitochondrial genome may be associated with clinical variability in HD, this suggestion was not supported by our preliminary experiments that examined the DdeI mitochondrial restriction fragment length polymorphism at position 10,394. Excitotoxicity has been a favored mechanism to explain the cell death in HD, particularly since intrastriatal injection of excitatory amino acids in animals creates HD-like pathology. Accordingly, we investigated the GluR6 kainate receptor. Of the variance in the age of onset of HD that was not accounted for by the CAG repeats, 13% could be attributed to GluR6 genotype variation. These data implicate GluR6-mediated excitotoxicity in the pathogenesis of HD and highlight the potential importance of this process in other polyglutamine repeat expansion diseases.
Keywords: excitotoxicity
Huntington disease (HD) is an autosomal dominant, progressive, neurodegenerative condition that is associated with abnormally large CAG repeat expansions in exon 1 of the IT15 gene localized on chromosome 4p. The CAG repeats are translated into a polyglutamine tract. Normal alleles with fewer than 36 uninterrupted repeats are generally stable, whereas disease alleles have both germ-line and somatic instability of repeat number, with a tendency for expansion in male transmissions. Since increasing length of the CAG repeats is related to decreasing age of onset of symptoms, the disease tends to present at earlier ages in successive generations, a process called anticipation. It is not clear how the resulting gene products with expanded polyglutamine tracts result in the region-specific cell death seen in this disease (1).
A number of studies have shown that the CAG repeat number on HD disease chromosomes only accounts for 50–61% of the variance of the age of onset (2–4). The contribution of the CAG repeat number to age of onset is less marked at the lower end of the disease size range and accounts for about 20% of the variance in individuals with fewer than 52 repeats (3). The poor predictive value in this size range is exemplified by our findings of cases with 36–39 repeats who have remained without a diagnosis of HD into their 80s and 90s (5).
Kremer et al. (6) analyzed CAG repeat length on disease chromosomes in late-onset HD cases and argued that there are familial factors distinct from the CAG repeat size that influence the age of onset of HD. Accordingly, we assembled a panel of 293 patients whose ages of onset were known and determined the relationship between CAG repeat size and age of onset of disease. After allowing for this association, we tested for possible genotype–age-of-onset association effects at the following loci by using a multiple linear regression approach:
(i) Variation associated with the normal allele of the HD gene has been associated with the age of onset of HD by Farrer et al. (7), who found a significant tendency for sibs who shared the same age of onset to inherit the same D4S10 allele from the normal parent. Snell et al. (2) reported a relationship between the number of CAG repeats on the normal allele and the age of onset of HD when the disease was maternally inherited. This model is compatible with the data of Farrer et al. (7), if one argues that the disease chromosome is less likely to undergo large expansions when maternally inherited and that paternally inherited alleles are more likely to have larger repeat numbers.
(ii) It is also possible that variation associated with the disease chromosome that lies outside the CAG repeats may affect the age of onset of disease. The Δ2642 polymorphism in the HD gene has two alleles (8). The insertion allele codes for four glutamines starting at codon 2642 and the deletion allele codes for three glutamines. The deletion allele is found on about 7% of normal chromosomes but on about 38% of disease chromosomes. Thus, 89% of HD patients heterozygous for the Δ2642 polymorphism will carry the deletion on the disease chromosome (38% × 93%/[38% × 93% + 62% × 7%]). This disequilibrium allows one to examine any putative functional consequences on the age of onset of HD of this codon-loss polymorphism when associated with HD chromosomes.
(iii) Apolipoprotein E (apoE) has recently attracted much interest, since allelic variation at this locus modifies the age of onset of Alzheimer disease. This gene has three common alleles. The apoE4 allele is associated with earliest age of onset, the E3 allele is associated with an intermediate age of onset, and the E2 allele is associated with the latest age of onset (9). The pathophysiology of this process is unclear. However, this locus may also exert modifying effects on a number of other distinct neurological diseases. ApoE allelic variation has been reported to be associated with risk of Lewy Body disease (10), age of onset of Creutzfeld–Jacob disease (11), bulbar symptoms in motor neurone disease (12), poor response after head injury (13), and even schizophrenia (14).
(iv) The role of mitochondrial metabolism in HD patients has been suggested by observations of decreased oxygen and glucose metabolism in the basal ganglia and cerebral cortex and postmortem findings of defects in mitochondrial metabolism and decreased complex 1 activity in platelets (15, 16). Beal and colleagues (16) have suggested that mitochondrial DNA sequence variants may interact with the HD mutation and affect the age of onset of HD. Since the mitochondrial genome is essentially linked, we have examined a panel of patients by using the comparatively ancient DdeI restriction fragment length polymorphism (RFLP) at position 10,934 (17, 18).
(v) Excitotoxicity has long been considered to be a potential mechanism that can account for the patterns of cell death seen in HD, since intrastriatal injections of excitatory amino acids in animals produce similar striatal pathology and neurochemistry to that seen in HD. Excitatory amino acids are abundant in the striatum, and binding sites for these neurotransmitters are reduced in HD brains. These data led to suggestions that their receptors were directly involved in the pathogenesis of HD (1). The GluR6 kainate receptor binds to excitatory amino acids and maps to human chromosome 6 (19). The polymorphic TAA repeat in its 3′ untranslated region (19) was used to determine if this locus was involved in modulating the age of onset of HD.
METHODS
Two hundred ninety-three patients with clinical diagnoses of HD whose ages of onset were available were ascertained from the Genetics Clinics in Cambridge, Manchester, and Oxford.
The CAG repeat sizes and the Δ2642 polymorphism in the HD gene were determined as described (20). ApoE genotypes (21), the mitochondrial 10,934 DdeI RFLP (18), and the TAA repeats in GluR6 (19) were determined as described.
The linear dependence of age of onset of HD on CAG repeat number was determined by linear regression and the goodness of fit was assessed by the proportion of variation in the dependent variable (age) explained by the regression line (the coefficient of determination, R2). The best fit for our data was obtained with the logarithmic transformation [log(age) = α + β(CAG repeat number)] (see below). Residuals from this model were checked and there was no evidence of departure from normality and equality of variance assumptions (results not shown). The possible genotypic effects of the five polymorphisms were assessed with multiple linear regression, while allowing for the predictive effects of the CAG repeat size. Although the CAG repeat number on the HD chromosome was considered as a numerical variable, we had no reason to consider any of the putative modifying loci as numerical variables, except for the CAG repeat number on the normal chromosomes (which was also considered as a nominal variable). Thus, all of the other putative modifying genotypes were only considered as nominal variables. This analysis was validated by using a permutation argument (see Results). Statistical analysis was performed with s-plus.
RESULTS
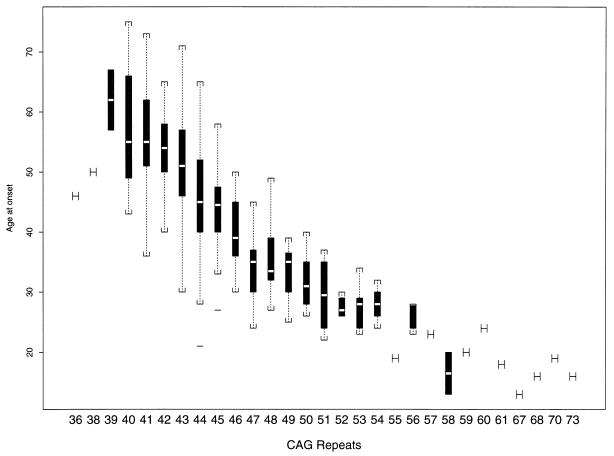
The relationship between age of onset of HD and CAG repeat size on disease chromosomes was determined by using linear regression for the entire sample (Fig. 1). As with earlier studies (2–4), the age of onset is linearly related to the number of CAG repeats (R2 = 0.59, P < 0.0001). However, a better fit for the data was obtained by using the logarithmic transformation: log(age) = α + β(CAG repeat number), where α = 6.15 (SE = 0.095) and β = −0.053 (SE = 0.0021). This parsimonious model resulted in the CAG repeats with a higher predictive value of 69% for the variation in the age of onset of HD (R2 = 0.693, P < 0.0001).
Figure 1.
Relationship between CAG repeat number on HD disease chromosome and age of onset of disease. For each repeat size, the median value is indicated as an open bar, the 95% confidence interval is indicated as the solid box, the range is indicated by brackets, and outlying points by solid lines.
For analyses of possible additional modifying genes for the age of onset of HD, we used a multiple linear regression approach. R2 values were determined for genotypes at each individual locus in conjunction with the influence of the CAG repeats on HD chromosomes. This value was compared with the R2 value for the CAG repeats alone, to determine the change in R2 (ΔR2) attributable to each putative modifying locus. These data and the significance of the ΔR2 for each of the candidate modifying loci are shown in Table 1. The CAG repeat length on normal chromosomes, Δ2642 genotypes, apoE genotypes, and the mitochondrial DdeI RFLP at position 10,394 failed to show any association with the age of onset of HD (Table 1).
Table 1.
Multiple linear regression analysis of possible candidate genes affecting the age of onset of HD, which are distinct from the CAG repeat number on HD chromosomes
| Model | R2 | ΔR2 | % unexplained variance | P value |
|---|---|---|---|---|
| HD CAG | 0.6933 | — | — | 0.0001 |
| HD CAG + normal CAG (numerical) | 0.681 | −0.012 | — | 0.061 |
| HD CAG + normal CAG (nominal) | 0.6848 | −0.0085 | — | 0.85 |
| HD CAG + apoE | 0.6999 | 0.0066 | 2.15 | 0.516 |
| HD CAG + mitochondrial DdeI site | 0.6959 | 0.0026 | 0.88 | 0.183 |
| HD CAG + Δ2642 genotype | 0.6934 | 0.0001 | 0.03 | 0.90 |
| HD CAG + GluR6 genotype | 0.734 | 0.41 | 13.36 | 0.008 |
Variance in the age of onset for the HD chromosome’s CAG repeats is indicated alone, as well as in combination with the different loci that were examined. ΔR2 indicates the relative improvement of the regression model when the genotypes at each of the candidate modifier loci are added to the HD CAG repeats. The percent unexplained variance indicates the percentage of the variance that could not be accounted for by the HD CAGs that were attributable to the candidate modifier loci. The CAG repeat number on HD chromosomes was considered as a numerical variable. The CAG repeat number on the normal chromosome was considered as both a nominal and a numerical variable. In both models, the ΔR2 value was a negative value, suggesting that the normal CAG repeat number worsened the fit of the data when added to the CAG repeat data on the disease chromosomes. In both cases, the P value was not significant. All of the other putative modifying loci were considered as nominal variables only.
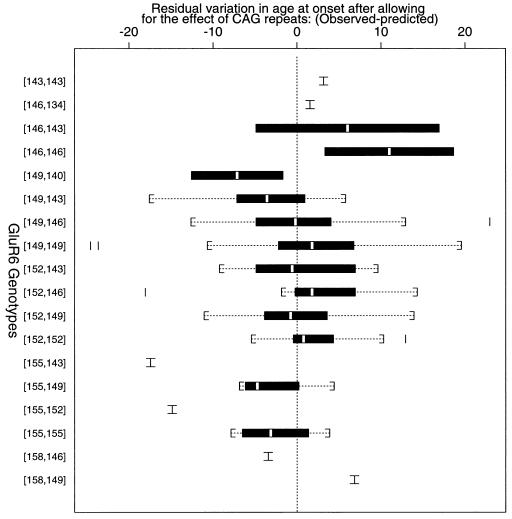
Genotypes at the GluR6 kainate receptor locus were determined by using primers that amplify a polymorphic TAA repeat in the 3′ untranslated portion of the gene (19). The addition of this locus to the CAG repeat number in the multiple linear regression approach resulted in a significant improvement to the fit and the R2 value increased from 69.3% to 73.4% (P = 0.008, Table 1). Therefore, 4.1% of the total variance can be attributed to GluR6 genotype variation, representing 13% of the variance in the age of onset of HD that was not accounted for by the CAG repeats (31%). Fig. 2 shows the association of different GluR6 genotypes with the residual variation in the age of onset of HD, after the effect of the CAG repeats have been accounted for. Since this is a regression analysis (and not a χ2 analysis), the results should not be affected by the small sample sizes associated with certain GluR6 genotypes.
Figure 2.
Relationship of GluR6 genotype to the amount of residual variation of the age of onset of HD, after the HD CAG repeats have been accounted for. Expected age of onset was determined by using the formula: log(age) = α + β(CAG repeat number), where α = 6.15 and β = −0.053. For each genotype, the median value is indicated as an open bar, the 95% confidence interval is indicated as the solid box, the range is indicated by brackets, and outlying points by solid lines. Genotypes are indicated by allele sizes in base pairs. Genotypes and the corresponding number of cases are as follows: 143,143, 1 case; 146,134, 1 case; 146,143, 2 cases; 146,146, 2 cases; 149,140, 2 cases; 149,143, 17 cases; 149,146, 30 cases; 149,149, 129 cases; 152,143, 7 cases; 152,146, 9 cases; 152,149, 56 cases; 152,152, 10 cases; 155,143, 1 case; 155,149, 4 cases; 155,152, 1 case; 155,155, 4 cases; 158,146, 1 case; 158,149, 1 case.
In addition to the regression analysis, we have also confirmed the significance of our results by using a permutation argument, which does not use distributional assumptions. This is an empirical method, where, after keeping CAG repeat number and age of onset fixed, each of the 293 patients was randomly assigned 1 of the 18 observed GluR6 genotypes. For each random assignment, an appropriate statistic (F statistic) was computed while allowing for the effect of the CAG repeats alone on the age of onset of HD. In 10,000 random permutations, only 120 random samples gave evidence of association equal to or more extreme than our observed experimental data (P = 0.012). Thus, this analysis provides independent support for an association of GluR6 genotypes with differing ages of onset of HD.
Furthermore, we used a more conservative strategy to test for the effects of the GluR6 locus on the age of onset of HD: Permutation analyses (using 10,000 random permutations) also showed significant effects for the GluR6 locus on the age of onset of HD even when the data were confined to patients who had GluR6 genotypes represented more than once (P = 0.025) or patients with 48 or fewer CAG repeats whose GluR6 genotypes were represented more than once (P = 0.019).
DISCUSSION
In this study, an expression that related the logarithm of the age of onset of HD to a function of the CAG repeat number accounted for almost 70% of the variance in the age of onset of HD. Thus, about 30% of the variation in the age of onset in HD patients is not explained by the HD chromosomes’ CAG repeat length. It is likely that some of this variance is associated with the difficulty in determining an exact age of onset in a disease that has an insidious presentation.
Most of the previous large studies (e.g., refs. 2–4) that have investigated this relationship used PCR assays that measured both the CAG repeats and the adjacent polymorphic CCG repeats, which can themselves vary by up to eight trinucleotide lengths (5). We used an assay that specifically measures CAG repeat length. Perhaps this can account for the slightly greater predictive value of the CAG repeat number for age of onset of disease in our sample.
The magnitude of the effect of other familial risk factors (other than the size of the CAG repeats) on the age of onset of HD has not been experimentally determined, although Kremer et al. (6) have presented and reviewed data that argue for the existence of such effects. One approach would be to examine HD families with multiple affected members and then compute the variance in the age of onset, with and without correction for the CAG repeat size. The magnitude of the familial effects can be determined by comparing the familial variance to the population variance, after correcting for repeat size. This type of approach has been used for the related CAG/polyglutamine expansion disease, spinocerebellar ataxia type 1, where Ranum et al. (22) have shown that interfamilial differences independent of repeat size explain about 5% of the age-of-onset variation.
CAG repeat number on the normal allele of HD patients did not affect the age of onset, and thus, a functional effect of the polyglutamine tract length in the normal protein on HD pathogenesis is unlikely. The Δ2642 polymorphism in the HD gene is associated with three or four glutamines starting at this position. Previous studies have not considered that this coding variant may affect the disease. The comparison of insertion homozygotes with heterozygotes is strongly biased toward comparing genotypes on disease chromosomes, since 89% of heterozygotes with HD are likely to have the deletion associated with the disease chromosome (see Introduction). Our data suggest that the deletion is unlikely to exert any effect when associated with disease chromosomes. The deletion on normal chromosomes is also unlikely to impact on HD, since the ages of onset of the homozygote deletion cases (n = 12) were similar to the other groups.
Our preliminary analysis of the mitochondrial 10,394 DdeI RFLP does not provide support for the hypothesis that mitochondrial DNA variants influence the course of HD. This polymorphism is likely to be old, since it is associated with an ancient deep split in the mitochondrial phylogenetic tree (17) and is informative in the population of the United Kingdom. Thus, more recent putative functional variants would be associated with one of the RFLP alleles. However, this limited analysis will need to be extended to a more detailed mitochondrial haplotype comparison in a larger set of patients before this model can be confidently ruled out.
The polymorphisms that are being analyzed in apoE correspond to arginine–cysteine interchanges at positions 112 and 158 that are thought to be the functional variants that impact upon the risk of Alzheimer disease (9). Although this locus may affect a large number of seemingly unrelated neuropsychiatric diseases, it does not affect the age of onset of HD.
Two different statistical analyses of our data showed significant associations of genotypes at the GluR6 locus with variation in the age of onset of HD. Although it is most likely that the TAA repeats are acting as a neutral polymorphism in linkage disequilibrium with a functional variant in the GluR6 gene, we cannot exclude the theoretical possibility of linkage disequilibrium with a variant in another nearby gene. Thus, we believe that these results implicate GluR6-mediated excitotoxicity in the pathogenesis of HD. The kainate-preferring GluR6 receptor is expressed in the cerebral cortex (19). Wagster et al. (23) showed specific losses of kainate and α-amino-3-hydroxy-5-methyl-4-isoxasole propionic acid (AMPA) binding in layer VI of the cerebral cortex of HD patients but no loss of N-methylaspartate binding. Since this layer of the cerebral cortex is associated with neuronal loss in HD, it has been postulated that the possession of kainate receptors may be a factor that can account for the selective vulnerability of specific neurones in HD (23). It is also possible that excitotoxic processes are occurring in other neurodegenerative diseases associated with polyglutamine expansions.
Of the variance in the age of onset of HD that was not accounted for by the CAG repeats, 13% could be attributed to GluR6 genotype variation. Although the effect associated with the TAA genotypes at this locus on the age of onset of HD is comparatively small, it may be masking a larger effect, because of the nature of the TAA repeats. Our study has revealed at least nine alleles and it is likely that this trinucleotide repeat shows appreciable mutation rates over time compared with diallelic polymorphisms. If the TAA repeats are acting as a neutral marker in linkage disequilibrium with a nearby functional variant, then this linkage disequilibrium will be eroded over time, because the repeats will mutate up and down and alleles that are of identical size will often not be identical by descent. The power to detect effects is also compromised since we have had to measure effects as a function of genotypes, as opposed to looking at the consequences of having particular alleles.
The likelihood that many monogenic diseases may be complex and influenced by a number of genetic and environmental processes is exemplified by studies in mice that have invoked modifying genes for cystic fibrosis (24). The presence of a modifying gene in HD in humans strengthens the argument of Estivill (25) that the boundaries between monogenic and multifactorial diseases may often be blurred. The discovery of even minor modifying effects can be of importance, since this allows the delineation of processes involved in disease pathogenesis, some which may be targets for therapeutic intervention.
Acknowledgments
We thank the many clinicians who have been involved with the HD patients and Doug Easton for discussions about the statistical analysis. This work was supported by the Huntington’s Disease Association, United Kingdom, and the Rehabilitation and Medical Research Trust (D.C.R.).
ABBREVIATIONS
- HD
Huntington disease
- apoE
apolipoprotein E
- RFLP
restriction fragment length polymorphism
References
- 1.Hayden M R, Kremer B. In: The Metabolic Bases of Inherited Disease. Scriver C R, Beaudet A L, Sly W S, Valle D, editors. New York: McGraw–Hill; 1995. pp. 4483–4510. [Google Scholar]
- 2.Snell R G, Macmillan J C, Cheadle J P, Fenton I, Lazarou L P, Davies P, MacDonald M E, Gusella J F, Harper P S, Shaw D J. Nat Genet. 1993;4:393–397. doi: 10.1038/ng0893-393. [DOI] [PubMed] [Google Scholar]
- 3.Duyao M, Ambrose C, Myers R, Novelletto A, Persichetti F, et al. Nat Genet. 1993;4:387–392. doi: 10.1038/ng0893-387. [DOI] [PubMed] [Google Scholar]
- 4.Andrew S E, Goldberg Y P, Kremer B, Telenius H, Theilmann J, Adam S, Starr E, Squitieri F, Lin B, Kalchmann M A, Graham R K, Hayden M R. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 5.Rubinsztein D C, Leggo J, Coles R, Almqvist E, Biancalana V, et al. Am J Hum Genet. 1996;56:16–22. [PMC free article] [PubMed] [Google Scholar]
- 6.Kremer B, Squitieri F, Telenius H, Andrew S, Theilmann J, Spence N, Goldberg Y P, Hayden M R. J Med Genet. 1993;30:991–995. doi: 10.1136/jmg.30.12.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Farrer L A, Cupples L A, Wiater P, Conneally P M, Gusella J F, Myers R H. Am J Hum Genet. 1993;53:125–130. [PMC free article] [PubMed] [Google Scholar]
- 8.Ambrose C M, Duyao M P, Barnes G, Bates G P, Lin C S, et al. Somatic Cell Mol Genet. 1994;20:27–38. doi: 10.1007/BF02257483. [DOI] [PubMed] [Google Scholar]
- 9.Hardy J. Am J Med Genet. 1995;60:456–460. doi: 10.1002/ajmg.1320600519. [DOI] [PubMed] [Google Scholar]
- 10.Pickering-Brown S M, Mann D M A, Bourke J P, Roberts D A, Balderson D, Burns A, Byrne J, Owen F. Lancet. 1994;343:1155. doi: 10.1016/s0140-6736(94)90257-7. [DOI] [PubMed] [Google Scholar]
- 11.Pickering-Brown S M, Mann D M A, Owen F, Ironside J W, De Siva R, Roberts D A, Balderson D J, Cooper P N. Neurosci Lett. 1995;187:127–129. doi: 10.1016/0304-3940(95)11353-3. [DOI] [PubMed] [Google Scholar]
- 12.Al-Chalabi A, Enayat Z E, Bakker M C, Sham P C, Ball D M, Shaw C E, Lloyd C M, Powell J F, Leigh P N. Lancet. 1996;347:159–160. doi: 10.1016/s0140-6736(96)90343-8. [DOI] [PubMed] [Google Scholar]
- 13.Nicoll J A R, Roberts G W, Graham D I. Nat Med. 1995;1:135–137. doi: 10.1038/nm0295-135. [DOI] [PubMed] [Google Scholar]
- 14.Harrington C R, Roth M, Xuereb J H, McKenna P J, Wischik C M. Neurosci Lett. 1995;202:101–104. doi: 10.1016/0304-3940(95)12218-4. [DOI] [PubMed] [Google Scholar]
- 15.Beal M F, Hyman B T, Koroshetz W. Trends Neurosci. 1993;16:125–131. doi: 10.1016/0166-2236(93)90117-5. [DOI] [PubMed] [Google Scholar]
- 16.Jenkins B G, Koroshetz W J, Beal M F, Rose B R. Neurology. 1993;43:2689–2695. doi: 10.1212/wnl.43.12.2689. [DOI] [PubMed] [Google Scholar]
- 17.Wallace D C. Am J Hum Genet. 1994;57:201–223. [PMC free article] [PubMed] [Google Scholar]
- 18.Schoffner J M, Brown M D, Torroni A, Lott M T, Cabell M F, Miorra S S, Beal M F, Yang C-C, Gearing M, Salvo R, Watts R L, Juncos J L, Hansen L A, Crain B J, Fayad M, Reckord C L, Wallace D C. Genomics. 1993;17:171–184. doi: 10.1006/geno.1993.1299. [DOI] [PubMed] [Google Scholar]
- 19.Paschen W, Blackstone C D, Huganir R L, Ross C A. Genomics. 1994;20:435–440. doi: 10.1006/geno.1994.1198. [DOI] [PubMed] [Google Scholar]
- 20.Rubinsztein D C, Leggo J, Goodburn S, Barton D E, Ferguson-Smith M A. Hum Mol Genet. 1995;5:203–206. doi: 10.1093/hmg/4.2.203. [DOI] [PubMed] [Google Scholar]
- 21.Hixson J E, Vernier D T. J Lipid Res. 1990;31:545–548. [PubMed] [Google Scholar]
- 22.Ranum L P W, Chung M-Y, Banfi S, Bryer A, Schut L J, Ramesar R, Duvick L A, McCall A, Subramony S H, Goldfarb L, Gomez C, Sandkuijl L A, Orr H T, Zoghbi H Y. Am J Hum Genet. 1994;55:244–252. [PMC free article] [PubMed] [Google Scholar]
- 23.Wagster M V, Hedreen J C, Peyser C E, Folstein S E, Ross C A. Exp Neurol. 1994;127:70–75. doi: 10.1006/exnr.1994.1081. [DOI] [PubMed] [Google Scholar]
- 24.Rozmahel R, Wichanski M, Matin A, Plyte S, Oliver M, Auerbach W, Moore A, Forstner J, Durie P, Nadeau J, Bear C, Tsui L C. Nat Genet. 1996;12:280–287. doi: 10.1038/ng0396-280. [DOI] [PubMed] [Google Scholar]
- 25.Estivill X. Nat Genet. 1996;12:348–350. doi: 10.1038/ng0496-348. [DOI] [PubMed] [Google Scholar]