Abstract
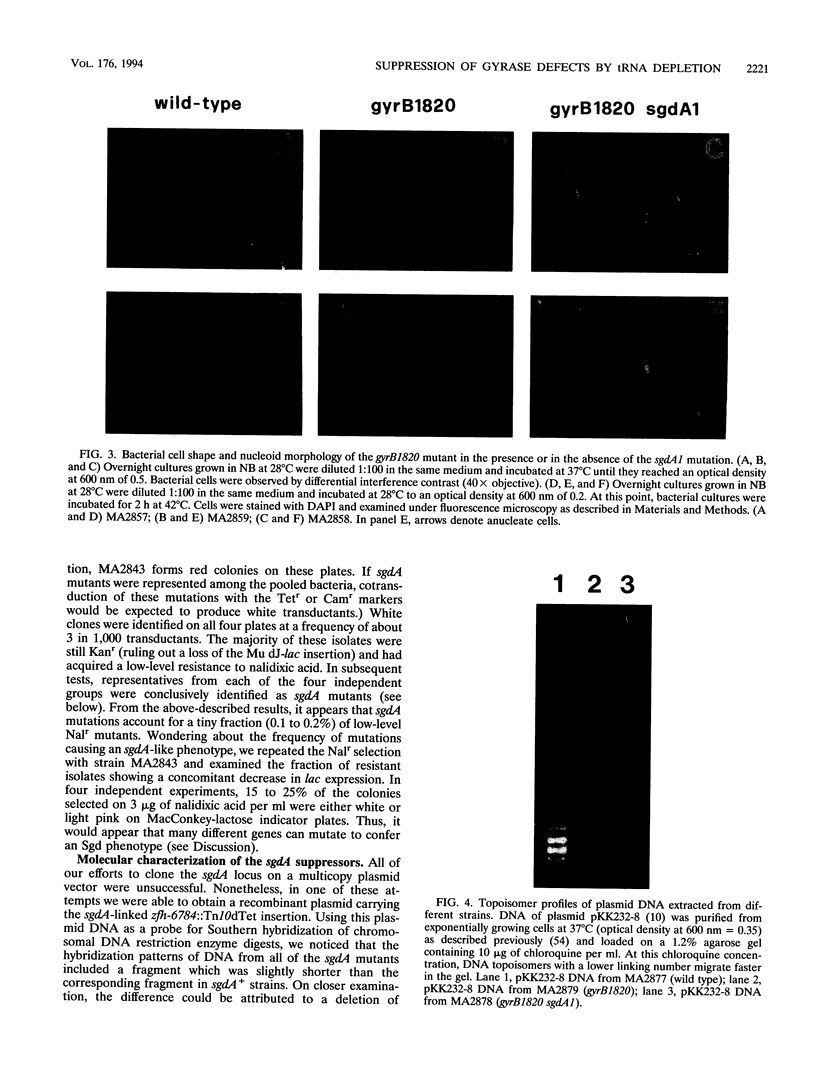
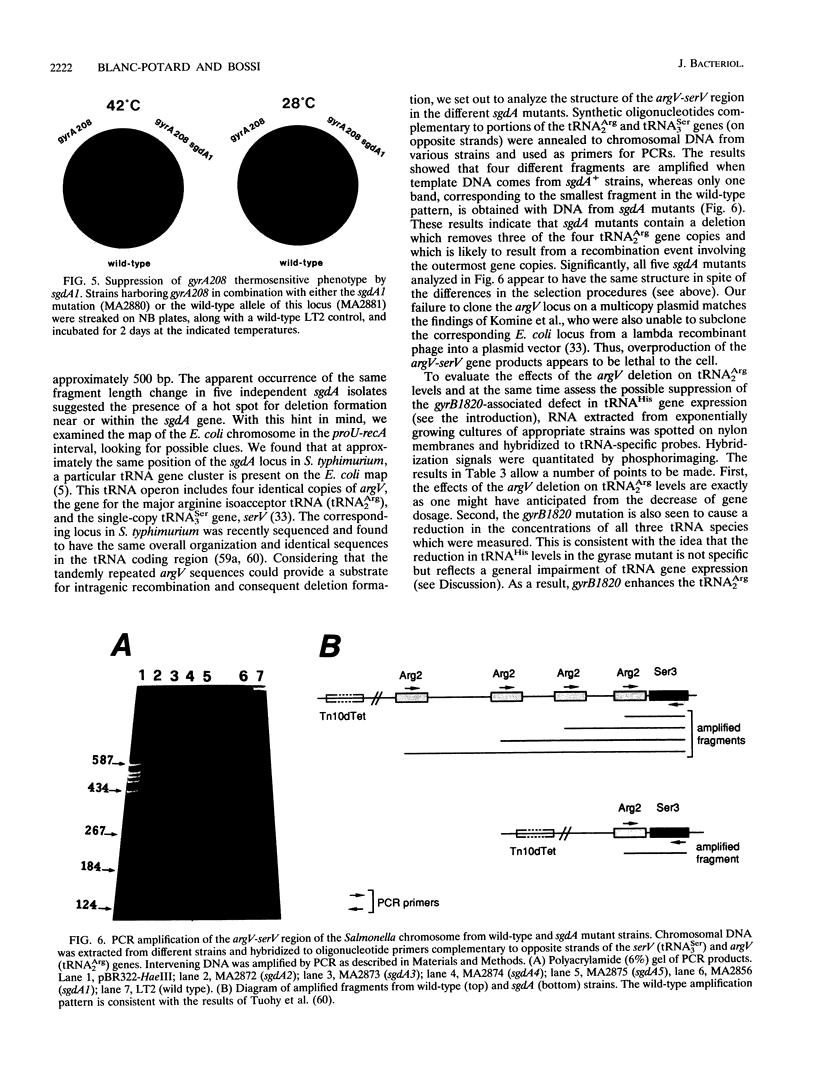
One of the pleiotropic phenotypes of mutations affecting DNA gyrase activity in Salmonella typhimurium is the constitutive deattenuation of the histidine operon. In the present work, we isolated and characterized a suppressor mutation which restores his attenuation in the presence of a defective gyrase. Such a suppressor, initially named sgdA1 (for suppressor gyrase deficiency), was found to correct additional phenotypes associated with defective gyrase function. These include the aberrant nucleoid partitioning of a gyrB mutant and the conditional lethality of a gyrA mutation. Furthermore, the sgdA1 mutation was found to confer low-level resistance to nalidixic acid. The last phenotype permitted isolation of a number of additional sgdA mutants. Genetic analysis established the recessive character of these alleles as well as the position of the sgdA locus at 57 U on the Salmonella genetic map. All of the sgdA mutants result from the same molecular event: a deletion removing three of the four tandemly repeated copies of argV, the gene which specifies tRNA(2Arg), the major arginine isoacceptor tRNA. These findings, combined with the observation of some Sgd-like phenotypes in a tRNA modification mutant (hisT mutant), lead us to propose that protein synthesis contributes, directly or indirectly, to the pathology of gyrase alterations in growing bacteria. We discuss plausible mechanisms which may be responsible for these effects.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams D. E., Bliska J. B., Cozzarelli N. R. Cre-lox recombination in Escherichia coli cells. Mechanistic differences from the in vitro reaction. J Mol Biol. 1992 Aug 5;226(3):661–673. doi: 10.1016/0022-2836(92)90623-r. [DOI] [PubMed] [Google Scholar]
- Adams D. E., Shekhtman E. M., Zechiedrich E. L., Schmid M. B., Cozzarelli N. R. The role of topoisomerase IV in partitioning bacterial replicons and the structure of catenated intermediates in DNA replication. Cell. 1992 Oct 16;71(2):277–288. doi: 10.1016/0092-8674(92)90356-h. [DOI] [PubMed] [Google Scholar]
- Antón D. N. Histidine regulatory mutants in Salmonella typhimurium. V. Two new classes histidine regulatory mutants. J Mol Biol. 1968 May 14;33(3):533–546. doi: 10.1016/0022-2836(68)90304-5. [DOI] [PubMed] [Google Scholar]
- Aota S., Gojobori T., Ishibashi F., Maruyama T., Ikemura T. Codon usage tabulated from the GenBank Genetic Sequence Data. Nucleic Acids Res. 1988;16 (Suppl):r315–r402. doi: 10.1093/nar/16.suppl.r315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachmann B. J. Linkage map of Escherichia coli K-12, edition 8. Microbiol Rev. 1990 Jun;54(2):130–197. doi: 10.1128/mr.54.2.130-197.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker T. A., Funnell B. E., Kornberg A. Helicase action of dnaB protein during replication from the Escherichia coli chromosomal origin in vitro. J Biol Chem. 1987 May 15;262(14):6877–6885. [PubMed] [Google Scholar]
- Bliska J. B., Cozzarelli N. R. Use of site-specific recombination as a probe of DNA structure and metabolism in vivo. J Mol Biol. 1987 Mar 20;194(2):205–218. doi: 10.1016/0022-2836(87)90369-x. [DOI] [PubMed] [Google Scholar]
- Bossi L., Kohno T., Roth J. R. Genetic characterization of the sufj frameshift suppressor in Salmonella typhimurium. Genetics. 1983 Jan;103(1):31–42. doi: 10.1093/genetics/103.1.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bossi L., Smith D. M. Conformational change in the DNA associated with an unusual promoter mutation in a tRNA operon of Salmonella. Cell. 1984 Dec;39(3 Pt 2):643–652. doi: 10.1016/0092-8674(84)90471-9. [DOI] [PubMed] [Google Scholar]
- Brosius J. Plasmid vectors for the selection of promoters. Gene. 1984 Feb;27(2):151–160. doi: 10.1016/0378-1119(84)90136-7. [DOI] [PubMed] [Google Scholar]
- Chen M. X., Bouquin N., Norris V., Casarégola S., Séror S. J., Holland I. B. A single base change in the acceptor stem of tRNA(3Leu) confers resistance upon Escherichia coli to the calmodulin inhibitor, 48/80. EMBO J. 1991 Oct;10(10):3113–3122. doi: 10.1002/j.1460-2075.1991.tb07865.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condemine G., Smith C. L. Transcription regulates oxolinic acid-induced DNA gyrase cleavage at specific sites on the E. coli chromosome. Nucleic Acids Res. 1990 Dec 25;18(24):7389–7396. doi: 10.1093/nar/18.24.7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka L. N. A third L-proline permease in Salmonella typhimurium which functions in media of elevated osmotic strength. J Bacteriol. 1982 Sep;151(3):1433–1443. doi: 10.1128/jb.151.3.1433-1443.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiNardo S., Voelkel K. A., Sternglanz R., Reynolds A. E., Wright A. Escherichia coli DNA topoisomerase I mutants have compensatory mutations in DNA gyrase genes. Cell. 1982 Nov;31(1):43–51. doi: 10.1016/0092-8674(82)90403-2. [DOI] [PubMed] [Google Scholar]
- Drlica K. Biology of bacterial deoxyribonucleic acid topoisomerases. Microbiol Rev. 1984 Dec;48(4):273–289. doi: 10.1128/mr.48.4.273-289.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Figueroa N., Wills N., Bossi L. Common sequence determinants of the response of a prokaryotic promoter to DNA bending and supercoiling. EMBO J. 1991 Apr;10(4):941–949. doi: 10.1002/j.1460-2075.1991.tb08028.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Filutowicz M., Jonczyk P. The gyrB gene product functions in both initiation and chain polymerization of Escherichia coli chromosome replication: suppression of the initiation deficiency in gyrB-ts mutants by a class of rpoB mutations. Mol Gen Genet. 1983;191(2):282–287. doi: 10.1007/BF00334827. [DOI] [PubMed] [Google Scholar]
- Garcia G. M., Mar P. K., Mullin D. A., Walker J. R., Prather N. E. The E. coli dnaY gene encodes an arginine transfer RNA. Cell. 1986 May 9;45(3):453–459. doi: 10.1016/0092-8674(86)90331-4. [DOI] [PubMed] [Google Scholar]
- Gellert M. DNA topoisomerases. Annu Rev Biochem. 1981;50:879–910. doi: 10.1146/annurev.bi.50.070181.004311. [DOI] [PubMed] [Google Scholar]
- Gellert M., Mizuuchi K., O'Dea M. H., Nash H. A. DNA gyrase: an enzyme that introduces superhelical turns into DNA. Proc Natl Acad Sci U S A. 1976 Nov;73(11):3872–3876. doi: 10.1073/pnas.73.11.3872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grosjean H., Fiers W. Preferential codon usage in prokaryotic genes: the optimal codon-anticodon interaction energy and the selective codon usage in efficiently expressed genes. Gene. 1982 Jun;18(3):199–209. doi: 10.1016/0378-1119(82)90157-3. [DOI] [PubMed] [Google Scholar]
- Hane M. W., Wood T. H. Escherichia coli K-12 mutants resistant to nalidixic acid: genetic mapping and dominance studies. J Bacteriol. 1969 Jul;99(1):238–241. doi: 10.1128/jb.99.1.238-241.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitzler J., Maréchal-Drouard L., Dirheimer G., Keith G. Use of a dot blot hybridization method for identification of pure tRNA species on different membranes. Biochim Biophys Acta. 1992 Feb 11;1129(3):273–277. doi: 10.1016/0167-4781(92)90503-r. [DOI] [PubMed] [Google Scholar]
- Hiraga S., Niki H., Ogura T., Ichinose C., Mori H., Ezaki B., Jaffé A. Chromosome partitioning in Escherichia coli: novel mutants producing anucleate cells. J Bacteriol. 1989 Mar;171(3):1496–1505. doi: 10.1128/jb.171.3.1496-1505.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Conditionally transposition-defective derivative of Mu d1(Amp Lac). J Bacteriol. 1984 Jul;159(1):130–137. doi: 10.1128/jb.159.1.130-137.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussain K., Elliott E. J., Salmond G. P. The parD- mutant of Escherichia coli also carries a gyrAam mutation. The complete sequence of gyrA. Mol Microbiol. 1987 Nov;1(3):259–273. doi: 10.1111/j.1365-2958.1987.tb01932.x. [DOI] [PubMed] [Google Scholar]
- Ishii S., Murakami T., Shishido K. Gyrase inhibitors increase the content of knotted DNA species of plasmid pBR322 in Escherichia coli. J Bacteriol. 1991 Sep;173(17):5551–5553. doi: 10.1128/jb.173.17.5551-5553.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnston H. M., Barnes W. M., Chumley F. G., Bossi L., Roth J. R. Model for regulation of the histidine operon of Salmonella. Proc Natl Acad Sci U S A. 1980 Jan;77(1):508–512. doi: 10.1073/pnas.77.1.508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Imamura R., Niki H., Hiraga S., Suzuki H. New topoisomerase essential for chromosome segregation in E. coli. Cell. 1990 Oct 19;63(2):393–404. doi: 10.1016/0092-8674(90)90172-b. [DOI] [PubMed] [Google Scholar]
- Kato J., Nishimura Y., Suzuki H. Escherichia coli parA is an allele of the gyrB gene. Mol Gen Genet. 1989 May;217(1):178–181. doi: 10.1007/BF00330959. [DOI] [PubMed] [Google Scholar]
- Kleckner N., Bender J., Gottesman S. Uses of transposons with emphasis on Tn10. Methods Enzymol. 1991;204:139–180. doi: 10.1016/0076-6879(91)04009-d. [DOI] [PubMed] [Google Scholar]
- Komine Y., Adachi T., Inokuchi H., Ozeki H. Genomic organization and physical mapping of the transfer RNA genes in Escherichia coli K12. J Mol Biol. 1990 Apr 20;212(4):579–598. doi: 10.1016/0022-2836(90)90224-A. [DOI] [PubMed] [Google Scholar]
- Koo H. S., Wu H. Y., Liu L. F. Effects of transcription and translation on gyrase-mediated DNA cleavage in Escherichia coli. J Biol Chem. 1990 Jul 25;265(21):12300–12305. [PubMed] [Google Scholar]
- Leclerc G., Sirard C., Drapeau G. R. The Escherichia coli cell division mutation ftsM1 is in serU. J Bacteriol. 1989 Apr;171(4):2090–2095. doi: 10.1128/jb.171.4.2090-2095.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis J. A., Ames B. N. Histidine regulation in Salmonella typhimurium. XI. The percentage of transfer RNA His charged in vivo and its relation to the repression of the histidine operon. J Mol Biol. 1972 Apr 28;66(1):131–142. doi: 10.1016/s0022-2836(72)80011-1. [DOI] [PubMed] [Google Scholar]
- Liu L. F., Wang J. C. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci U S A. 1987 Oct;84(20):7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lockshon D., Morris D. R. Positively supercoiled plasmid DNA is produced by treatment of Escherichia coli with DNA gyrase inhibitors. Nucleic Acids Res. 1983 May 25;11(10):2999–3017. doi: 10.1093/nar/11.10.2999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lodge J. K., Kazic T., Berg D. E. Formation of supercoiling domains in plasmid pBR322. J Bacteriol. 1989 Apr;171(4):2181–2187. doi: 10.1128/jb.171.4.2181-2187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luttinger A. L., Springer A. L., Schmid M. B. A cluster of genes that affects nucleoid segregation in Salmonella typhimurium. New Biol. 1991 Jul;3(7):687–697. [PubMed] [Google Scholar]
- Lynch A. S., Wang J. C. Anchoring of DNA to the bacterial cytoplasmic membrane through cotranscriptional synthesis of polypeptides encoding membrane proteins or proteins for export: a mechanism of plasmid hypernegative supercoiling in mutants deficient in DNA topoisomerase I. J Bacteriol. 1993 Mar;175(6):1645–1655. doi: 10.1128/jb.175.6.1645-1655.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murray M. L., Hartman P. E. Overproduction of hisH and hisF gene products leads to inhibition of cell cell division in Salmonella. Can J Microbiol. 1972 May;18(5):671–681. doi: 10.1139/m72-105. [DOI] [PubMed] [Google Scholar]
- Norris V., Alliotte T., Jaffé A., D'Ari R. DNA replication termination in Escherichia coli parB (a dnaG allele), parA, and gyrB mutants affected in DNA distribution. J Bacteriol. 1986 Nov;168(2):494–504. doi: 10.1128/jb.168.2.494-504.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Byrne C. P., Ní Bhriain N., Dorman C. J. The DNA supercoiling-sensitive expression of the Salmonella typhimurium his operon requires the his attenuator and is modulated by anaerobiosis and by osmolarity. Mol Microbiol. 1992 Sep;6(17):2467–2476. doi: 10.1111/j.1365-2958.1992.tb01423.x. [DOI] [PubMed] [Google Scholar]
- Orr E., Fairweather N. F., Holland I. B., Pritchard R. H. Isolation and characterisation of a strain carrying a conditional lethal mutation in the cou gene of Escherichia coli K12. Mol Gen Genet. 1979;177(1):103–112. doi: 10.1007/BF00267259. [DOI] [PubMed] [Google Scholar]
- Palmer D. T., Blum P. H., Artz S. W. Effects of the hisT mutation of Salmonella typhimurium on translation elongation rate. J Bacteriol. 1983 Jan;153(1):357–363. doi: 10.1128/jb.153.1.357-363.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pruss G. J., Manes S. H., Drlica K. Escherichia coli DNA topoisomerase I mutants: increased supercoiling is corrected by mutations near gyrase genes. Cell. 1982 Nov;31(1):35–42. doi: 10.1016/0092-8674(82)90402-0. [DOI] [PubMed] [Google Scholar]
- Richardson S. M., Higgins C. F., Lilley D. M. The genetic control of DNA supercoiling in Salmonella typhimurium. EMBO J. 1984 Aug;3(8):1745–1752. doi: 10.1002/j.1460-2075.1984.tb02041.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roth J. R., Antón D. N., Hartman P. E. Histidine regulatory mutants in Salmonella typhimurium. I. Isolation and general properties. J Mol Biol. 1966 Dec 28;22(2):305–323. doi: 10.1016/0022-2836(66)90134-3. [DOI] [PubMed] [Google Scholar]
- Rudd K. E., Menzel R. his operons of Escherichia coli and Salmonella typhimurium are regulated by DNA supercoiling. Proc Natl Acad Sci U S A. 1987 Jan;84(2):517–521. doi: 10.1073/pnas.84.2.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanderson K. E., Roth J. R. Linkage map of Salmonella typhimurium, edition VII. Microbiol Rev. 1988 Dec;52(4):485–532. doi: 10.1128/mr.52.4.485-532.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spirito F., Figueroa-Bossi N., Bossi L. The relative contributions of transcription and translation to plasmid DNA supercoiling in Salmonella typhimurium. Mol Microbiol. 1994 Jan;11(1):111–122. doi: 10.1111/j.1365-2958.1994.tb00294.x. [DOI] [PubMed] [Google Scholar]
- Steck T. R., Drlica K. Bacterial chromosome segregation: evidence for DNA gyrase involvement in decatenation. Cell. 1984 Apr;36(4):1081–1088. doi: 10.1016/0092-8674(84)90058-8. [DOI] [PubMed] [Google Scholar]
- Steck T. R., Pruss G. J., Manes S. H., Burg L., Drlica K. DNA supercoiling in gyrase mutants. J Bacteriol. 1984 May;158(2):397–403. doi: 10.1128/jb.158.2.397-403.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura F., Nishimura S., Ohki M. The E. coli divE mutation, which differentially inhibits synthesis of certain proteins, is in tRNASer1. EMBO J. 1984 May;3(5):1103–1107. doi: 10.1002/j.1460-2075.1984.tb01936.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toone W. M., Rudd K. E., Friesen J. D. Mutations causing aminotriazole resistance and temperature sensitivity reside in gyrB, which encodes the B subunit of DNA gyrase. J Bacteriol. 1992 Aug;174(16):5479–5481. doi: 10.1128/jb.174.16.5479-5481.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsao Y. P., Wu H. Y., Liu L. F. Transcription-driven supercoiling of DNA: direct biochemical evidence from in vitro studies. Cell. 1989 Jan 13;56(1):111–118. doi: 10.1016/0092-8674(89)90989-6. [DOI] [PubMed] [Google Scholar]
- Turnbough C. L., Jr, Neill R. J., Landsberg R., Ames B. N. Pseudouridylation of tRNAs and its role in regulation in Salmonella typhimurium. J Biol Chem. 1979 Jun 25;254(12):5111–5119. [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Wu H. Y., Shyy S. H., Wang J. C., Liu L. F. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988 May 6;53(3):433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]