Abstract
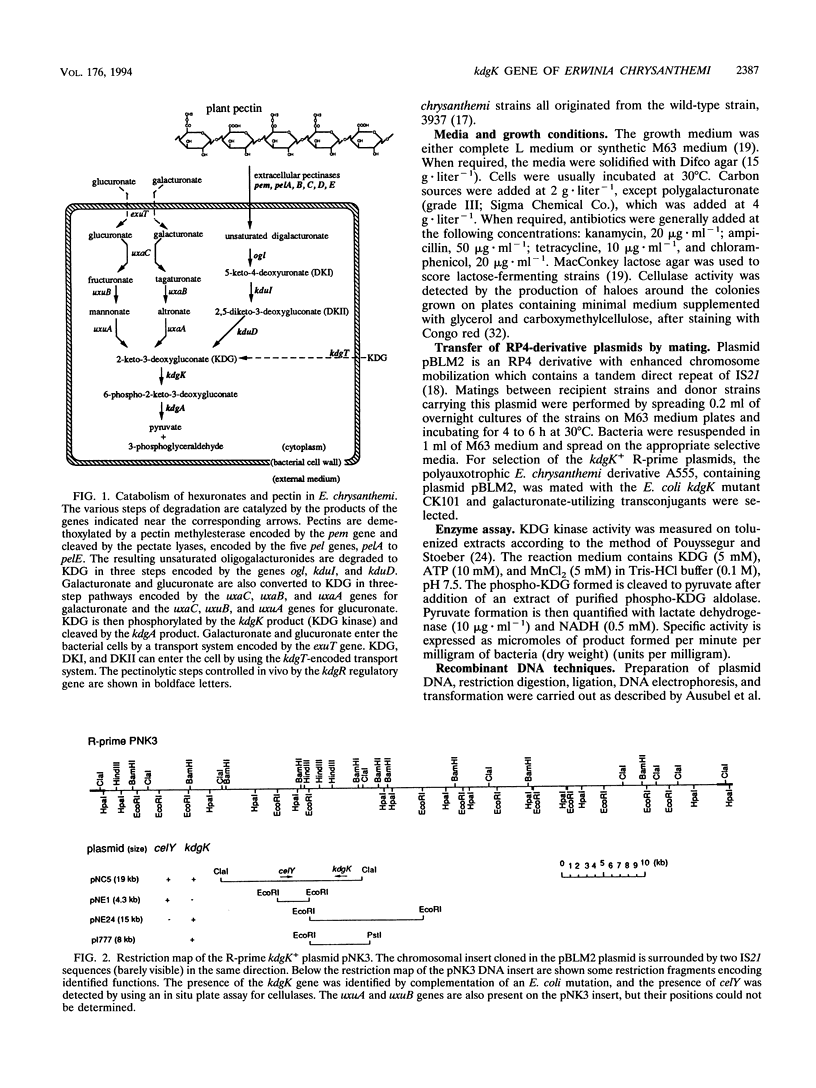
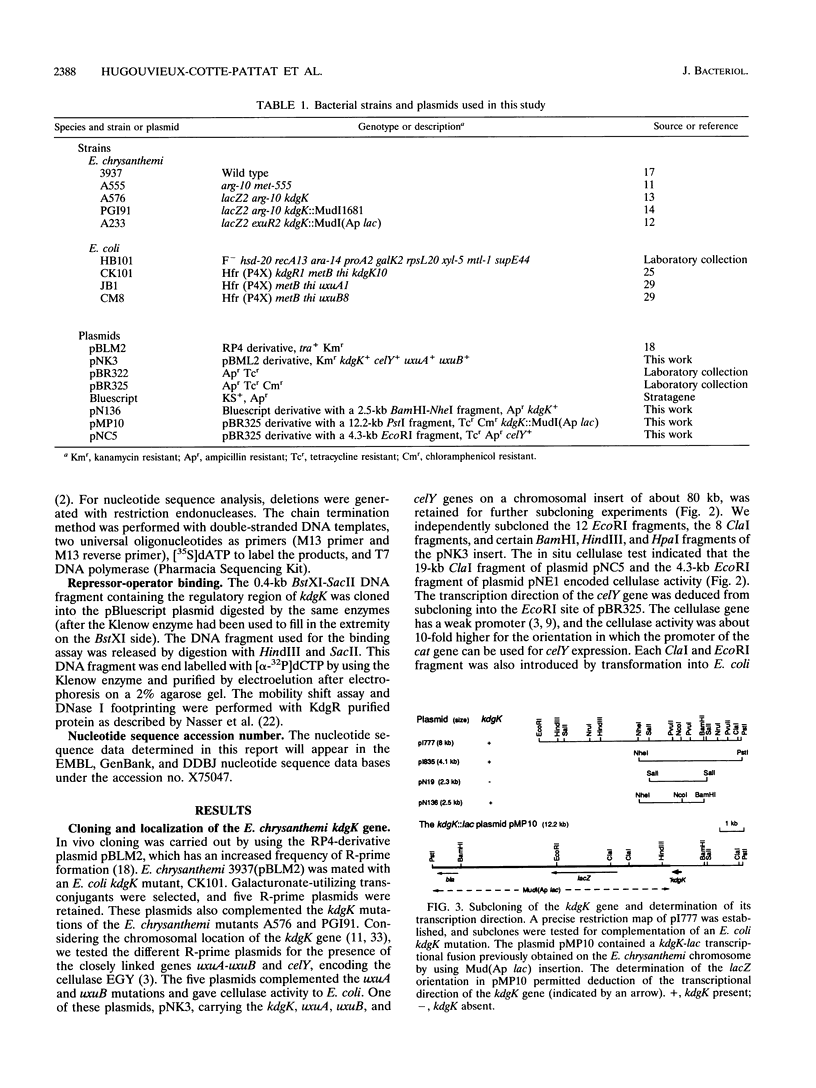
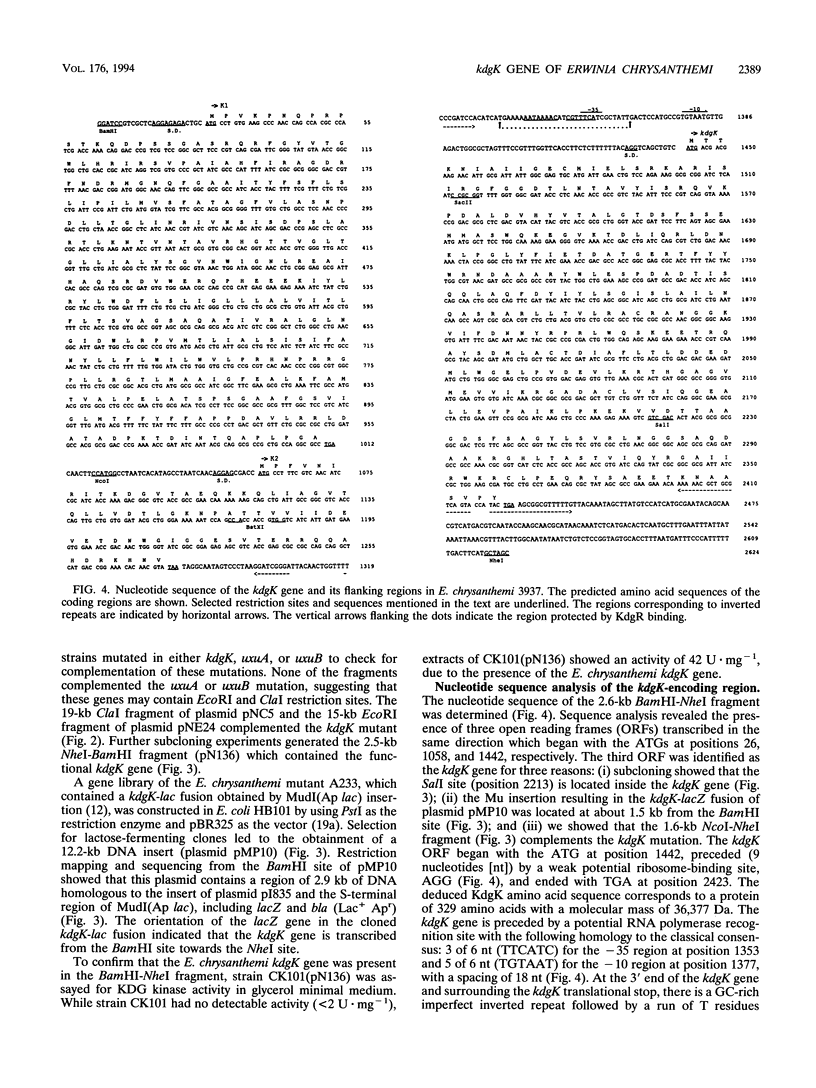
The pathways of pectin and galacturonate catabolism in Erwinia chrysanthemi converge to form a common intermediate, 2-keto-3-deoxygluconate (KDG), which is phosphorylated by KDG kinase encoded by the kdgK gene. We cloned the kdgK gene of E. chrysanthemi 3937 by complementing an Escherichia coli kdgK mutation, using an RP4-derivative plasmid. One of the kdgK R-prime plasmids harbored a DNA insert of about 80 kb and carried the uxuA and uxuB genes involved in glucuronate catabolism and the celY gene coding for an E. chrysanthemi cellulase. The kdgK and celY genes were precisely located on this plasmid, and their respective transcriptional directions were determined. The nucleotide sequence of the kdgK region indicated that the kdgK reading frame is 981 bases long, corresponding to a protein of 329 amino acids with a molecular mass of 36,377 Da. Analysis of the deduced primary amino acid sequence showed that this enzyme is a new member of the PfkB family of carbohydrate kinases. Expression of kdgK is controlled by a negative regulatory gene, kdgR, which represses all the steps of pectin degradation. Near the putative promoter of the kdgK gene, we identified a putative KdgR-binding site and demonstrated that the KdgR protein specifically binds in vitro to this DNA region. The KdgR-KDG couple directly mediates the phenomenon of repression or induction. The KDG kinase, by limiting the intracellular inducer concentration, appears to be a key enzyme in induction of the whole catabolic pathway.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aulkemeyer P., Ebner R., Heilenmann G., Jahreis K., Schmid K., Wrieden S., Lengeler J. W. Molecular analysis of two fructokinases involved in sucrose metabolism of enteric bacteria. Mol Microbiol. 1991 Dec;5(12):2913–2922. doi: 10.1111/j.1365-2958.1991.tb01851.x. [DOI] [PubMed] [Google Scholar]
- Aymeric J. L., Guiseppi A., Pascal M. C., Chippaux M. Mapping and regulation of the cel genes in Erwinia chrysanthemi. Mol Gen Genet. 1988 Jan;211(1):95–101. doi: 10.1007/BF00338398. [DOI] [PubMed] [Google Scholar]
- Chatterjee A. K., Thurn K. K., Tyrell D. J. Isolation and characterization of Tn5 insertion mutants of Erwinia chrysanthemi that are deficient in polygalacturonate catabolic enzymes oligogalacturonate lyase and 3-deoxy-D-glycero-2,5-hexodiulosonate dehydrogenase. J Bacteriol. 1985 May;162(2):708–714. doi: 10.1128/jb.162.2.708-714.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condemine G., Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Isolation of Erwinia chrysanthemi kduD mutants altered in pectin degradation. J Bacteriol. 1986 Mar;165(3):937–941. doi: 10.1128/jb.165.3.937-941.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Condemine G., Robert-Baudouy J. Analysis of an Erwinia chrysanthemi gene cluster involved in pectin degradation. Mol Microbiol. 1991 Sep;5(9):2191–2202. doi: 10.1111/j.1365-2958.1991.tb02149.x. [DOI] [PubMed] [Google Scholar]
- Guiseppi A., Aymeric J. L., Cami B., Barras F., Creuzet N. Sequence analysis of the cellulase-encoding celY gene of Erwinia chrysanthemi: a possible case of interspecies gene transfer. Gene. 1991 Sep 30;106(1):109–114. doi: 10.1016/0378-1119(91)90573-t. [DOI] [PubMed] [Google Scholar]
- Henikoff S., Henikoff J. G. Automated assembly of protein blocks for database searching. Nucleic Acids Res. 1991 Dec 11;19(23):6565–6572. doi: 10.1093/nar/19.23.6565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Reverchon S., Robert-Baudouy J. Expanded linkage map of Erwinia chrysanthemi strain 3937. Mol Microbiol. 1989 May;3(5):573–581. doi: 10.1111/j.1365-2958.1989.tb00204.x. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Analysis of the regulation of the pelBC genes in Erwinia chrysanthemi 3937. Mol Microbiol. 1992 Aug;6(16):2363–2376. doi: 10.1111/j.1365-2958.1992.tb01411.x. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Hexuronate catabolism in Erwinia chrysanthemi. J Bacteriol. 1987 Mar;169(3):1223–1231. doi: 10.1128/jb.169.3.1223-1231.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Isolation of Erwinia chrysanthemi mutants altered in pectinolytic enzyme production. Mol Microbiol. 1989 Nov;3(11):1587–1597. doi: 10.1111/j.1365-2958.1989.tb00144.x. [DOI] [PubMed] [Google Scholar]
- Hugouvieux-Cotte-Pattat N., Robert-Baudouy J. Molecular analysis of the Erwinia chrysanthemi region containing the kdgA and zwf genes. Mol Microbiol. 1994 Jan;11(1):67–75. doi: 10.1111/j.1365-2958.1994.tb00290.x. [DOI] [PubMed] [Google Scholar]
- Kotoujansky A., Lemattre M., Boistard P. Utilization of a thermosensitive episome bearing transposon TN10 to isolate Hfr donor strains of Erwinia carotovora subsp. chrysanthemi. J Bacteriol. 1982 Apr;150(1):122–131. doi: 10.1128/jb.150.1.122-131.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marrs B. Mobilization of the genes for photosynthesis from Rhodopseudomonas capsulata by a promiscuous plasmid. J Bacteriol. 1981 Jun;146(3):1003–1012. doi: 10.1128/jb.146.3.1003-1012.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasser W., Condemine G., Plantier R., Anker D., Robert-Baudouy J. Inducing properties of analogs of 2-keto-3-deoxygluconate on the expression of pectinase genes of Erwinia chrysanthemi. FEMS Microbiol Lett. 1991 Jun 1;65(1):73–78. doi: 10.1016/0378-1097(91)90474-o. [DOI] [PubMed] [Google Scholar]
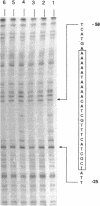
- Nasser W., Reverchon S., Condemine G., Robert-Baudouy J. Specific interactions of Erwinia chrysanthemi KdgR repressor with different operators of genes involved in pectinolysis. J Mol Biol. 1994 Feb 18;236(2):427–440. doi: 10.1006/jmbi.1994.1155. [DOI] [PubMed] [Google Scholar]
- Nasser W., Reverchon S., Robert-Baudouy J. Purification and functional characterization of the KdgR protein, a major repressor of pectinolysis genes of Erwinia chrysanthemi. Mol Microbiol. 1992 Jan;6(2):257–265. doi: 10.1111/j.1365-2958.1992.tb02007.x. [DOI] [PubMed] [Google Scholar]
- Portalier R., Robert-Baudouy J., Stoeber F. Regulation of Escherichia coli K-12 hexuronate system genes: exu regulon. J Bacteriol. 1980 Sep;143(3):1095–1107. doi: 10.1128/jb.143.3.1095-1107.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pouyssegur J., Stoeber F. Etude du rameu dégradatif commun des hexuronates chez Escherichia coli K 12. Purification, propriétés et individualité de la 2-céto-3-désoxy-D-gluconokinase. Biochimie. 1971;53(6):771–781. [PubMed] [Google Scholar]
- Pouyssegur J., Stoeber F. Genetic control of the 2-keto-3-deoxy-d-gluconate metabolism in Escherichia coli K-12: kdg regulon. J Bacteriol. 1974 Feb;117(2):641–651. doi: 10.1128/jb.117.2.641-651.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reverchon S., Huang Y., Bourson C., Robert-Baudouy J. Nucleotide sequences of the Erwinia chrysanthemi ogl and pelE genes negatively regulated by the kdgR gene product. Gene. 1989 Dec 21;85(1):125–134. doi: 10.1016/0378-1119(89)90472-1. [DOI] [PubMed] [Google Scholar]
- Reverchon S., Nasser W., Robert-Baudouy J. Characterization of kdgR, a gene of Erwinia chrysanthemi that regulates pectin degradation. Mol Microbiol. 1991 Sep;5(9):2203–2216. doi: 10.1111/j.1365-2958.1991.tb02150.x. [DOI] [PubMed] [Google Scholar]
- Robert-Baudouy J. M., Portalier R. C. Mutations affectant le catabolisme du glucuronate chez Escherichia coli K12. Mol Gen Genet. 1974;131(1):31–46. doi: 10.1007/BF00269385. [DOI] [PubMed] [Google Scholar]
- Shingler V., Powlowski J., Marklund U. Nucleotide sequence and functional analysis of the complete phenol/3,4-dimethylphenol catabolic pathway of Pseudomonas sp. strain CF600. J Bacteriol. 1992 Feb;174(3):711–724. doi: 10.1128/jb.174.3.711-724.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M., Wood P. J. Use of Congo red-polysaccharide interactions in enumeration and characterization of cellulolytic bacteria from the bovine rumen. Appl Environ Microbiol. 1982 Apr;43(4):777–780. doi: 10.1128/aem.43.4.777-780.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Gijsegem F., Toussaint A. In vivo cloning of Erwinia carotovora genes involved in the catabolism of hexuronates. J Bacteriol. 1983 Jun;154(3):1227–1235. doi: 10.1128/jb.154.3.1227-1235.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu L. F., Reizer A., Reizer J., Cai B., Tomich J. M., Saier M. H., Jr Nucleotide sequence of the Rhodobacter capsulatus fruK gene, which encodes fructose-1-phosphate kinase: evidence for a kinase superfamily including both phosphofructokinases of Escherichia coli. J Bacteriol. 1991 May;173(10):3117–3127. doi: 10.1128/jb.173.10.3117-3127.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Hippel P. H., Bear D. G., Morgan W. D., McSwiggen J. A. Protein-nucleic acid interactions in transcription: a molecular analysis. Annu Rev Biochem. 1984;53:389–446. doi: 10.1146/annurev.bi.53.070184.002133. [DOI] [PubMed] [Google Scholar]