Abstract
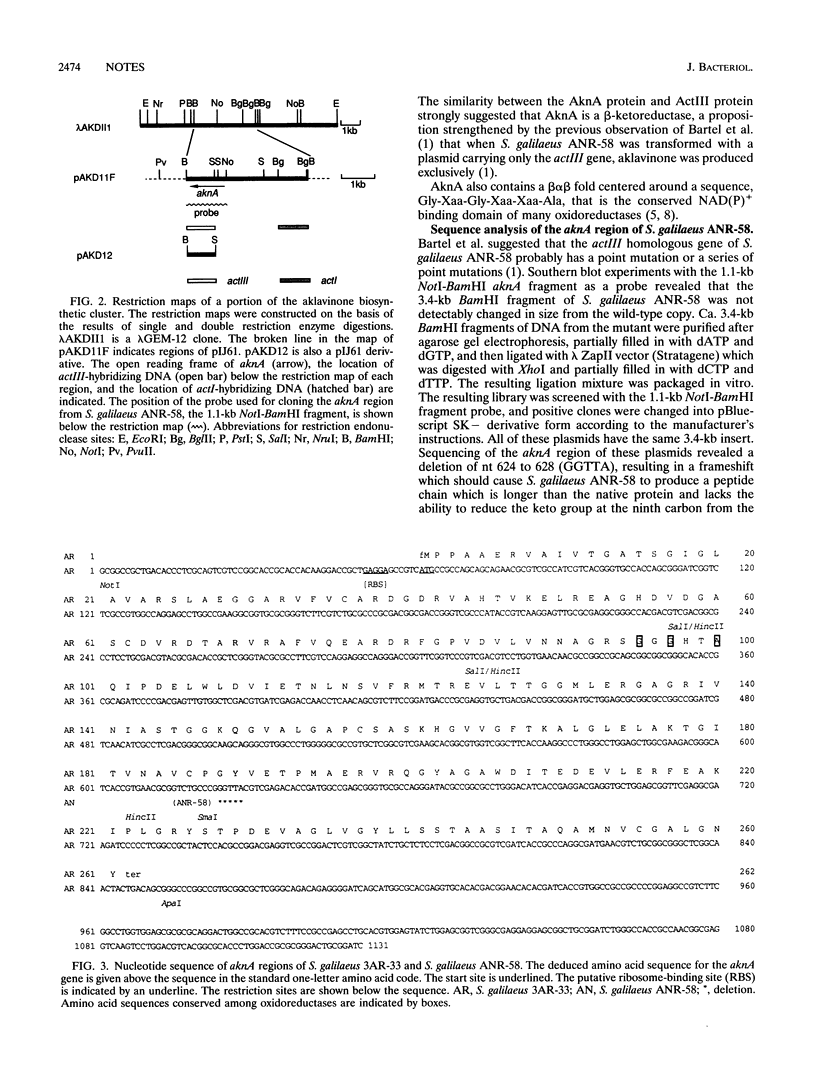
A 3.4-kb BamHI fragment that is assumed to be a part of the aklavinone biosynthetic gene cluster of Streptomyces galilaeus 3AR-33 and contains the genes required for the early stage of polyketide biosynthesis was sequenced. The nucleotide sequence of the region that hybridizes to the actIII probe reveals the presence of a gene, aknA, whose deduced protein product is very similar to the ActIII protein and other known oxidoreductases. The predicted AknA protein is believed to be responsible for catalyzing the reduction of the keto group at the ninth carbon from the carboxyl terminus of the assembled polyketide to the corresponding secondary alcohol. The predicted AknA protein has a calculated molecular mass of 27,197 Da (261 amino acids) and the highly conserved sequence Gly-Xaa-Gly-Xaa-Xaa-Ala commonly seen in oxidoreductases. Cloning and sequence analysis of the aknA region of the 2-hydroxyaklavinone-producing strain S. galilaeus ANR-58 identified an alteration in the gene, confirming that the aknA gene is essential for aklavinone biosynthesis.
Full text
PDF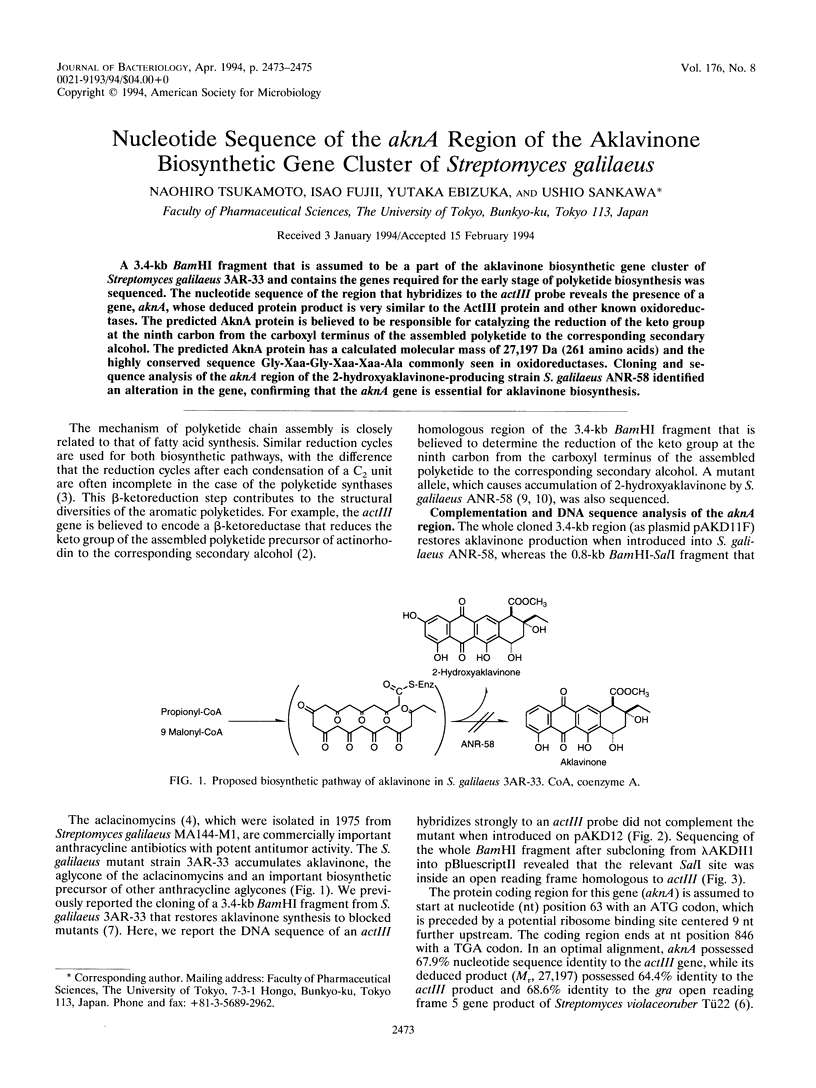

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bartel P. L., Zhu C. B., Lampel J. S., Dosch D. C., Connors N. C., Strohl W. R., Beale J. M., Jr, Floss H. G. Biosynthesis of anthraquinones by interspecies cloning of actinorhodin biosynthesis genes in streptomycetes: clarification of actinorhodin gene functions. J Bacteriol. 1990 Sep;172(9):4816–4826. doi: 10.1128/jb.172.9.4816-4826.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hallam S. E., Malpartida F., Hopwood D. A. Nucleotide sequence, transcription and deduced function of a gene involved in polyketide antibiotic synthesis in Streptomyces coelicolor. Gene. 1988 Dec 30;74(2):305–320. doi: 10.1016/0378-1119(88)90165-5. [DOI] [PubMed] [Google Scholar]
- Hopwood D. A., Sherman D. H. Molecular genetics of polyketides and its comparison to fatty acid biosynthesis. Annu Rev Genet. 1990;24:37–66. doi: 10.1146/annurev.ge.24.120190.000345. [DOI] [PubMed] [Google Scholar]
- Oki T., Kitamura I., Yoshimoto A., Matsuzawa Y., Shibamoto N., Ogasawara T., Inui T., Takamatsu A., Takeuchi T., Masuda T. Antitumor anthracycline antibiotics, aclacinomycin A and analogues. I. Taxonomy, production, isolation and physicochemical properties. J Antibiot (Tokyo) 1979 Aug;32(8):791–800. doi: 10.7164/antibiotics.32.791. [DOI] [PubMed] [Google Scholar]
- Sherman D. H., Malpartida F., Bibb M. J., Kieser H. M., Bibb M. J., Hopwood D. A. Structure and deduced function of the granaticin-producing polyketide synthase gene cluster of Streptomyces violaceoruber Tü22. EMBO J. 1989 Sep;8(9):2717–2725. doi: 10.1002/j.1460-2075.1989.tb08413.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsukamoto N., Fujii I., Ebizuka Y., Sankawa U. Cloning of aklavinone biosynthesis genes from Streptomyces galilaeus. J Antibiot (Tokyo) 1992 Aug;45(8):1286–1294. doi: 10.7164/antibiotics.45.1286. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A., Johdo O., Ishikura T., Takeuchi T. Anthracycline antibiotic 2-hydroxyaclacinomycins. I. 2-Hydroxyaclacinomycin-producing recombinant obtained from aclacinomycin-blocked mutants of Streptomyces galilaeus by a technique of protoplast fusion. Jpn J Antibiot. 1991 Mar;44(3):269–276. [PubMed] [Google Scholar]
- Yoshimoto A., Johdo O., Ishikura T., Takeuchi T. Anthracycline antibiotic 2-hydroxyaclacinomycins. II. Production of 2-hydroxyaclacinomycins A and B by a new recombinant strain and their antitumor activities. Jpn J Antibiot. 1991 Mar;44(3):277–286. [PubMed] [Google Scholar]



