Abstract
In the P53 tumor suppressor gene, a remarkably large number of somatic mutations are found at methylated CpG dinucleotides. We have previously mapped the distribution of (±) anti-7β,8α-dihydroxy-9α,10α-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE) adducts along the human P53 gene [Denissenko, M. F., Pao, A., Tang, M.-s. & Pfeifer, G. P. (1996) Science 274, 430–432]. Strong and selective formation of adducts occurred at guanines in CpG sequences of codons 157, 248, and 273, which are the major mutational hot spots in lung cancer. Chromatin structure was not involved in preferential modification of these sites by BPDE. To investigate other possible mechanisms underlying the selectivity of BPDE binding, we have mapped the adducts in plasmid DNA containing genomic P53 sequences. The adduct profile obtained was different from that in genomic DNA. However, when cytosines at CpG sequences were converted to 5-methylcytosines by the CpG-specific methylase SssI and the DNA was subsequently treated with BPDE, adduct hot spots were created which were similar to those seen in genomic DNA where all CpGs are methylated. A strong positive effect of 5-methylcytosine on BPDE adduct formation at CpG sites was also documented with sequences of the PGK1 gene derived from an active or inactive human X chromosome and having differential methylation patterns. These results show that methylated CpG dinucleotides, in addition to being an endogenous promutagenic factor, may represent a preferential target for exogenous chemical carcinogens. The data open new avenues concerning the reasons that the majority of mutational hot spots in human genes are at CpGs.
Keywords: benzo[a]pyrene, 5-methylcytosine
Mutational analysis of the P53 tumor suppressor gene provides a unique opportunity to investigate the etiology, epidemiology, and pathogenesis of human cancer (1–4). Close to 50% of all tumors are estimated to contain a mutation in P53 (5). Among all genetic alterations in P53, a remarkably high number of somatic mutations are found at methylated CpG dinucleotides. In fact, five major P53 mutational hot spots, i.e., codons 175, 245, 248, 273, and 282, contain methylated CpGs (6). Human tumors of different tissue origin display a different nature of inactivating mutations. Close to 50% of all colon cancers bear mutations at the three CpG hot spot codons 175, 248, and 273 (7). They are G → A transitions, thus implicating an endogenous methylation-driven process (most probably deamination of 5-methylcytosine; 5-mC) as a major causative factor. In contrast, about 25% of mutations are transitions at CpG sites in spontaneous breast carcinomas (1, 4), and only 10% of liver cancers contain such mutations (1). In total, the ratio of transitions to transversions is about 3:1 in colon tumors, 1:1 in breast tumors, and 1:3 in liver cancers (4). The percentage of transversions, especially of the G → T type, is high in lung tumors diagnosed in smokers (1). This type of cancer is characterized by three mutational hot spots at codons 157, 248, and 273 (4, 8), one of which (codon 157) is a common mutation site only in tumors of the lung. The predominant type of mutation found at these lung tumor hot spots is a G → T transversion. The majority of base substitutions in lung cancers can be ascribed to guanines on the nontranscribed DNA strand (1). Such peculiar tissue specificity of the P53 mutational spectrum strongly suggests that certain exogenous factors may be implicated as etiological agents in lung tumorigenesis. Of these agents, the potent environmental and tobacco smoke carcinogen benzo[a]pyrene (B[a]P) is one of the first to consider because of its distinct mutagenicity and the proven correlation between tobacco smoking and lung cancer (9). Upon metabolic activation, B[a]P is transformed to the ultimate carcinogenic compound (±) anti-7β,8α-dihydroxy-9α,10α-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene (BPDE), which generates in DNA predominantly covalent (+) trans adducts at the N2 position of guanine (10–13). These covalent adducts have been shown to be the basis for the mutagenic and carcinogenic effects of BPDE (14, 15). The genomic targets for a variety of chemical carcinogens may include oncogenes and tumor suppressor genes. Identification of a link between DNA damage and mutations will strengthen the understanding of the extent that elements of the environment are responsible for initiation of tumorigenesis in humans.
We have previously mapped the distribution of BPDE adducts along the human P53 gene (8). Strong and selective formation of adducts occurred at guanine positions in codons 157, 248, and 273. The pattern of BPDE–DNA adduction was nearly identical in three nonrelated cell types, including normal bronchial epithelial cells, but the basis for the specificity of adduct formation was unknown. Chromatin structure may be involved in determining the site-selectivity of carcinogen binding to DNA. Also, additional factors such as DNA sequence context or cytosine methylation patterns may participate in shaping the BPDE adduct profile in P53.
The epigenetic maintenance of 5-mC in DNA of higher eukaryotic organisms is indispensable for cell differentiation, X chromosome inactivation, and genomic imprinting (16). Recently, it became clear that DNA methylation may be a crucial factor in tumorigenesis (for reviews, see refs. 17–19). Events of local increases (20–22) or decreases (23, 24) in the extent of methylation were found in the human genome and correlated with tumor development. The hypermutability of CpG sequences has largely been attributed to spontaneous deamination of 5-mC to thymine causing a C → T transition mutation (17, 18). Thus, 5-mC-induced mutations have been considered as endogenous alterations mostly contributing to the background mutation rate (17).
In this work, we have attempted to assess the role of cytosine methylation in human carcinogenesis from a different angle. We have found that the presence of 5-mC within a CpG site has a strong positive effect on the reactivity of a CpG site with the carcinogen BPDE. These results show how CpG dinucleotides that are stably methylated in the human P53 gene in all tissues examined (6, 25, 26), in addition to being an endogenous promutagenic factor, may represent a preferential target for exogenous chemical carcinogens.
MATERIALS AND METHODS
Cell Culture and DNA Modification.
Human–hamster hybrid cells carrying either an active (cell line Y162-11C) or an inactive human X chromosome (cell line X8-6T2) were cultured as described (27), and genomic DNA was isolated according to standard procedures (28). HeLa S3 cells (obtained from the American Type Culture Collection) were grown under standard conditions. Cells were treated with BPDE as described (8). Plasmid pAT153P53π (29) was kindly provided by L. Crawford and S. P. Tuck (Imperial Cancer Research Fund, Cambridge, U.K.). It contains a genomic sequence of human P53 encompassing exons 2–11. Plasmid DNA was methylated in vitro using the CpG-specific methylase SssI (New England Biolabs) according to the manufacturer’s instructions. Control DNA was mock-methylated in the absence of S-adenosylmethionine (SAM). Completion of methylation was confirmed by digesting an aliquot of the reaction mixture with the methylation-sensitive restriction endonuclease HpaII and by Maxam–Gilbert sequencing. Racemic BPDE was purchased from the NCI repository (Midwest Research Institute, Kansas City, MO). Modification of DNA with BPDE was done according to published procedures (30, 31), which include repeated extractions of adducted DNA with organic solvents (water-saturated diethyl ether and isoamyl alcohol). Control DNA samples were treated with solvent (95% ethanol) only. Under these conditions, the achieved levels of modification were in the range of 4.5–51.8 adducts per 106 nucleotides for the respective carcinogen concentrations of 0.03–0.5 μM (32) or <1 adduct per fragment for the 336-bp 5′ end-labeled AvaII–SspI DNA fragment. The methods for DNA fragment isolation and 5′ end-labeling were the same as those previously described (33).
Treatment of DNA with UvrABC.
Purified DNA was treated with an excess of UvrABC (a 10-fold molar excess of protein over 104 nucleotides of DNA) as described (8, 34). Under the reaction conditions used, the cleavage at BPDE–DNA adducts by UvrABC nucleases is quantitative.
Ligation-Mediated PCR (LMPCR).
Oligonucleotide primers for LMPCR of the human P53 gene were described elsewhere (6, 35). A total of 250 pg of plasmid DNA was used in each reaction along with 1 μg of carrier Escherichia coli genomic DNA. Primer sets A and H were used to analyze the X chromosome-linked PGK1 gene (36, 37). LMPCR was done as described (38).
RESULTS
CpG Methylation Creates Hot Spots for BPDE Binding.
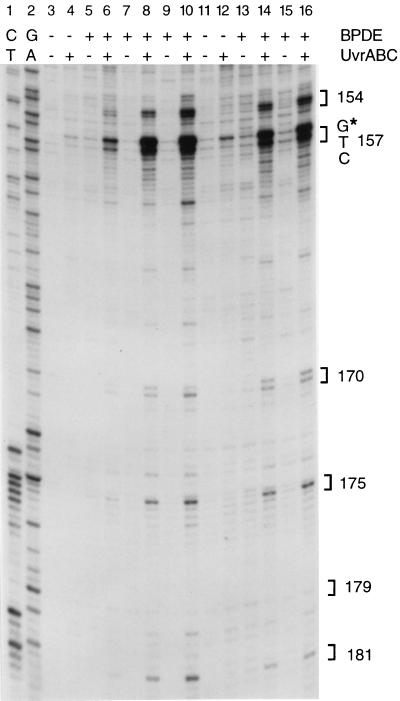
We have previously reported that BPDE–guanine adducts preferentially form at the major P53 mutational hot spots (codons 157, 248, and 273) in human lung cancers (8). Such selectivity of BPDE binding was not detected by Puisieux et al. (39) with BPDE-modified plasmid DNA containing P53 cDNA sequences. This difference suggests that a protein-associated chromatin structure which is absent in cloned DNA may affect BPDE adduct formation. We have therefore mapped the adducts in genomic DNA isolated from cells and subsequently treated with BPDE in vitro; we then compared the adduct pattern with that present in DNA from BPDE-treated cells. Purified genomic DNA was modified with BPDE and then reacted with UvrABC nucleases. UvrABC makes a dual incision seven nucleotides 5′ and four nucleotides 3′ to a BPDE adduct (30). The 3′ incisions can be precisely mapped by amplifying the resulting 5-phosphate-containing DNA using LMPCR with P53-specific primers (6, 8, 35). Fig. 1 shows that the profile of BPDE–DNA adduction in exon 5 is nearly identical in cells and in isolated DNA. The only significant difference is seen near the bottom of the gel, where Gs near codons 180 and 181 are protected from BPDE modification in cells (Fig. 1, lanes 11–16). This could be due to association of these sequences with a nucleosome core region (40) which may hinder BPDE adduct formation (41, 42). Nearly identical adduct patterns were seen in free DNA and in BPDE-treated cells along exons 7 and 8 (data not shown). This result rules out an involvement of chromatin structure as a major modulator in creating BPDE adduct hot spots in the P53 gene.
Figure 1.
Distribution of BPDE adducts along P53 exon 5. DNA (lanes 3–10) or cells (lanes 11–16) were treated with BPDE, and the distribution of adducts in P53 was determined after UvrABC incision and LMPCR. Concentrations of BPDE were 0.04 μM (lanes 5 and 6), 0.2 μM (lanes 7 and 8), 1 μM (lanes 9 and 10), 2 μM (lanes 13 and 14), and 4 μM (lanes 15 and 16). Lanes 1 and 2 are Maxam–Gilbert sequencing controls. The positions of P53 codons are indicated by brackets. The star indicates a mutational hot spot in codon 157.
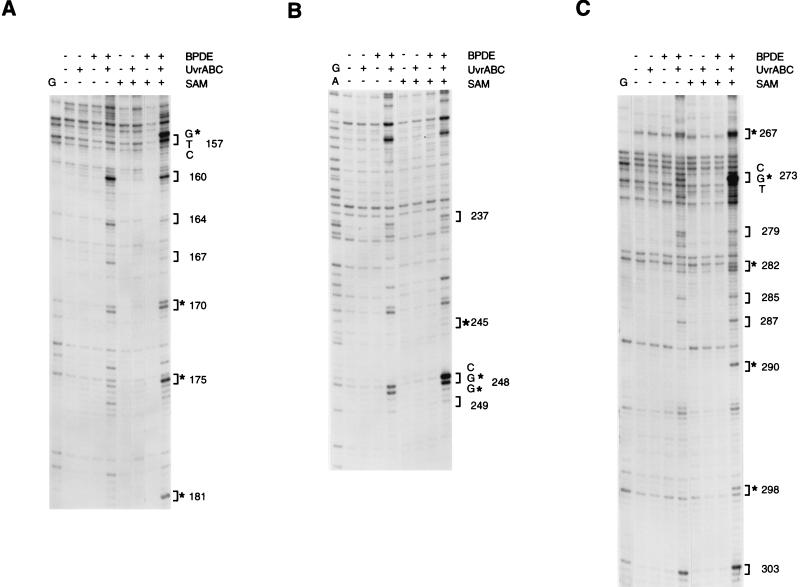
Another important factor that may determine the selectivity of BPDE binding is the DNA methylation pattern present in human genomic DNA. Notably, the CpG dinucleotides at the major mutational hot spots in the P53 coding region are known to be methylated in all human tissues examined (6, 25, 26). To test this hypothesis, we have determined the distribution of adducts in the nontranscribed strand of plasmid DNA containing genomic P53 sequences (Fig. 2). Since the plasmid DNA was isolated from E. coli, cytosines in this DNA are not methylated. The most significantly damaged bases in this nonmethylated DNA were guanines in codons 160, 164, 167, 170, and 174–176 (Fig. 2A) in exon 5; codons 237, 243, and 248 in exon 7 (Fig. 2B); and codons 267, 273, 275, 279, 285, 287, and 303 in exon 8 (Fig. 2C). The pattern of damaged bases in nonmethylated plasmid DNA lacked the damage hot spots seen previously with genomic DNA. However, when the plasmid DNA was methylated by using the CpG-specific methylase SssI and subsequently modified with BPDE, the adduct profile was quite dissimilar from that in nonmethylated DNA and very similar to that seen in genomic DNA (Fig. 2; compare with Fig. 1 and with figures 2 and 3 in ref. 8). In exon 5, strong damage hot spots included codons 156 and 157, and, to a lesser extent, codon 175. In exon 7, the strongest binding was documented at codon 248, which contained a HpaII site (CCGG). Here, the second guanine 3′ to 5-mC was damaged almost as much as the first guanine within the CpG site. Methylation of the CpG site in codon 248 was found to be ubiquitous and “tenacious” in human tissues (26). In exon 8, a prominent hot spot appeared at codon 273, and a lesser enhancement of binding occurred at codons 267, 282, 290, 298, and 303. In summary, the adduct distribution in methylated plasmid strikingly resembled the distribution obtained upon analysis of cells (8) or genomic DNA (Fig. 1). Almost all sites damaged preferentially in methylated DNA were guanines located in CpG sequences, although not all methylated CpG dinucleotides were affected equally. In other words, the extent of enhancement by methylation differed from site to site and was up to 10-fold. These results show that cytosine methylation is a critical modulating factor for BPDE binding to DNA.
Figure 2.
Distribution of BPDE adducts along P53 sequences in plasmid DNA differing in methylation status. DNA was methylated (+SAM) or mock-methylated (−SAM) with the CpG-specific methylase SssI, modified with BPDE (0.2 μM), and the distribution of adducts was analyzed by UvrABC incision and LMPCR. (A) Exon 5, nontranscribed strand. (B) Exon 7, nontranscribed strand. (C) Exon 8, nontranscribed strand. Stars mark codons containing CpG dinucleotides.
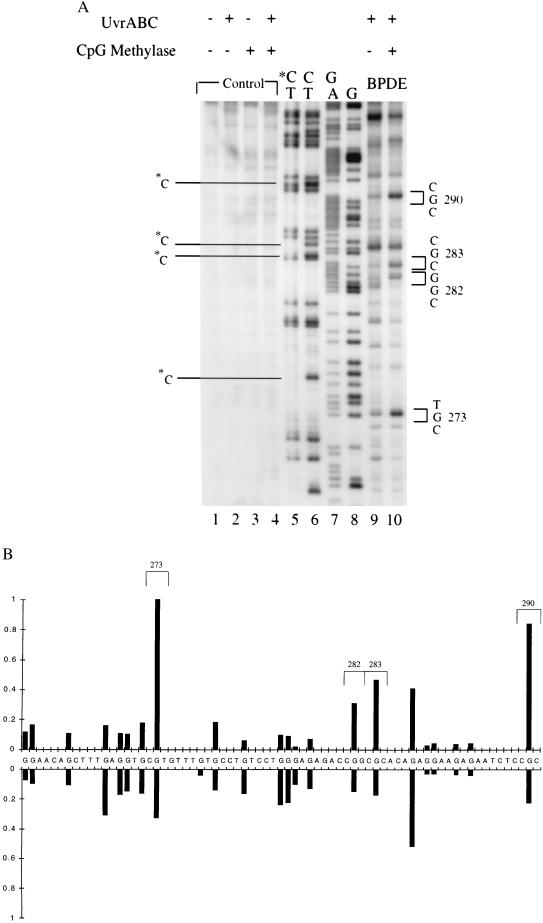
A similar result was obtained when adducted base positions were analyzed by the 5′ side incision reaction made by UvrABC nuclease on BPDE-modified substrates. UvrABC makes a dual incision seven nucleotides 5′ and four nucleotides 3′ to a BPDE adduct (30). The 5′ break positions, unlike the 3′ incisions, are not detectable by LMPCR analysis but can be detected by using 5′ end-labeled DNA fragments. An analysis of exon 8 shows that the reactivity of guanines with BPDE is selectively increased when cytosines in CpG sequences are converted to 5-mCs (Fig. 3). Similar results were obtained after 5′ end-labeling of fragments containing exons 5 and 7 (data not shown).
Figure 3.
Mapping of UvrABC-induced 5′ break positions in BPDE-modified methylated and unmethylated DNA fragments. A 5′ end-labeled AvaII–SspI DNA fragment containing sequences of exon 8 of the P53 gene was methylated with SssI or mock-methylated, treated with BPDE, and then reacted with UvrABC nucleases. (A) Autoradiogram. Lanes: 1–4, no BPDE treatment; 9 and 10, BPDE treatment of unmethylated and methylated DNA, respectively; 5–8, Maxam–Gilbert sequencing reactions. *C marks methylated cytosines at CpG sites indicated by a missing band in the C-specific reaction. (B) Quantitation. The intensities of BPDE adduct-induced UvrABC incisions at different sequences from methylated DNA (Upper) or unmethylated DNA (Lower) were quantitated by phosphorimaging.
BPDE Adduct Distribution in Human DNA Sequences Differing in Cytosine Methylation.
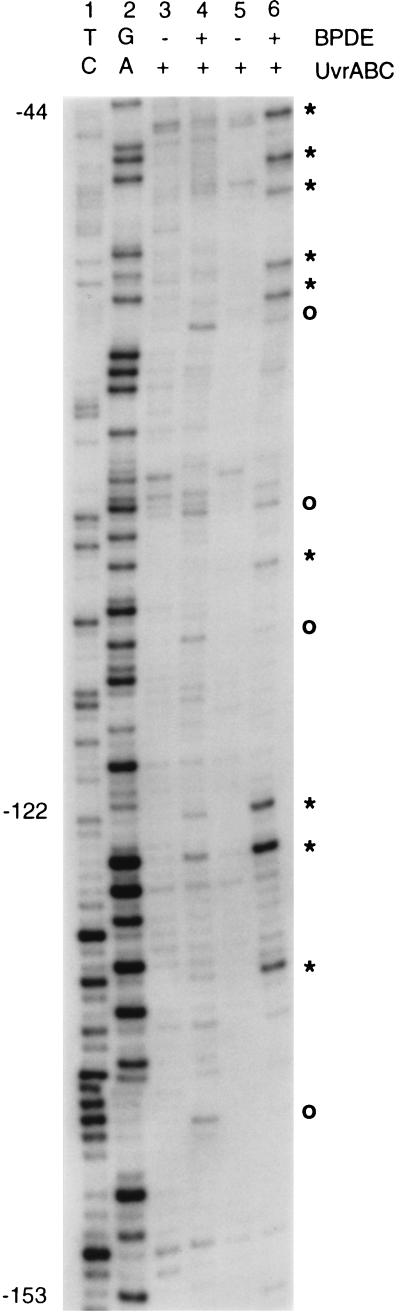
Results from the in vitro methylation study suggest that the density of CpG methylation might influence adduct formation. We have next analyzed identical DNA sequences in which a different methylation status is controlled epigenetically by a gene-silencing mechanism. The housekeeping genes located on the human X chromosome are subject to X inactivation. The promoter region of the X-linked PGK1 gene is a CpG island whose methylation status differs between the active X chromosome (Xa) and the inactive X chromosome (Xi). It was shown that the Xa in hamster–human hybrid cells is completely unmethylated at all 120 CpG sites located in this region. In contrast, 118 CpGs are methylated on the Xi (36, 37). We have treated DNA from hamster–human hybrid cell lines Y162-11C (Xa) and X8-6T2 (Xi) with BPDE and performed LMPCR using a primer set described earlier (36, 37). This primer set visualizes the regions of the PGK1 promoter where the methylation status is completely opposite in the Xi compared with the Xa. Fig. 4 shows a strong preference of BPDE adduct formation at methylated CpGs over their nonmethylated counterparts. With one exception, at all positions where the targeted G residue was immediately neighbored by a 5′ C, an apparent increase in BPDE reactivity with the Xi over the Xa was seen. Interestingly, the guanine base at position −122, which is preferentially modified in the Xi, is the second G in the sequence 5′-CGG-3′. This is very similar to the situation observed for the P53 codon 248 (Fig. 2B) and may be characteristic for this type of trinucleotide. All other sites showing increased BPDE binding in the methylated Xi DNA were at CpGs. The same tendency was documented with another primer set that spans another region of the CpG island located at the 5′ end of the PGK1 gene (data not shown). The data confirm that methylation of cytosine favors the interaction of an adjacent guanine with BPDE.
Figure 4.
Distribution of BPDE adducts along the methylated and unmethylated CpG island of the human PGK1 gene. DNA containing sequences from the Xa (unmethylated; lanes 3 and 4) or from the Xi (methylated; lanes 5 and 6) was treated with BPDE (1 μM), and the distribution of adducts in the PGK1 promoter and CpG island was determined after UvrABC incision and LMPCR. Lanes 1 and 2 are Maxam–Gilbert sequencing controls. Stars indicate positions where an increase of modification was noticed with the Xi DNA containing methylated PGK1 sequences; ○, other modified G positions.
DISCUSSION
The P53 mutational spectrum (i.e., the distribution of mutations along the gene) and the mutational signature (i.e., the characteristic ratio of transitions, transversions, deletions, etc.) is different between lung cancer and other cancer types, and the mutational signature also differs from germ-line mutations. In lung cancer, about 40% of the mutations are G → T transversions, most of them biased to a guanine on the nontranscribed DNA strand (1). In addition, there is a striking scarcity of transition mutations at CpG sequences in the P53 gene of lung cancer (9%). Transition mutations at CpG are much more frequent in almost all other cancers or in the germ line (up to 50%) and have been linked to deamination of endogenous 5-mC bases. Of the three mutational hot spots that are selectively damaged by BPDE (8), codon 157 is the hot spot unique to lung cancer. Mutations occur frequently at codons 248 and 273 also in other cancers, and are usually recovered there as transition mutations at CpG sequences (7). In lung cancers, however, G → T transversions predominate at these mutational hot spots. Although selection certainly plays a role in shaping the P53 mutational spectra in all cancers, there are many codons (about 140 in lung cancer and about 220 in all cancers combined) that can be targets of different missense mutations (7, 8). The striking coincidence of the lung cancer mutational spectrum in smokers and the B[a]P adduct spectrum (8), and the dominance of G → T transversion mutations in these cancers, have suggested that a large proportion of P53 mutations in lung cancer are not caused by endogenous processes but may be caused by a carcinogen of the polycyclic aromatic hydrocarbon class.
In this paper, we present evidence that cytosine methylation is the primary factor which directs the strongly preferential interaction of the ultimate carcinogenic compound BPDE with P53 DNA sequences. We show that the distribution of BPDE–DNA adducts differs drastically in CpG-methylated DNA compared with nonmethylated DNA. Moreover, the hot spots of BPDE binding in methylated plasmid DNA correspond exactly to the hot spots found in cells and in genomic DNA (8), whereas no such similarity is seen with nonmethylated DNA. Guanine residues in DNA are preferentially attacked by a variety of carcinogens, and BPDE is one of them. The effect of cytosine methylation of CpG sites on carcinogen-induced DNA modification has been analyzed in only a few other cases. N-methyl-N-nitrosourea-dependent modification of the N7 position of guanine was shown to be inhibited when 5-mC was a 5′ neighboring base (43). Methylation of cytosine also reduces formation of UV-induced (6–4) photoproducts (44, 45). However, formation of mitomycin C monoadducts and crosslinks is enhanced 1.4- to 3-fold by methylation of CpG sequences (46, 47).
How does cytosine methylation increase the reactivity of a CpG site with an electrophilic polycyclic aromatic hydrocarbon such as BPDE? The structural influence of the cytosine methyl group on B-DNA conformation seems to be insignificant as this group is located in the major groove of the DNA helix (48). In contrast, the B[a]P ring of (+)-trans adducts is positioned in the minor groove and is directed toward the 5′ end of the modified strand (49, 50). BPDE forms noncovalent intercalative complexes with double-stranded DNA that are thought to precede covalent binding of BPDE to DNA (51). It has previously been reported that methylation of the 5 position of cytosine gives rise to an enhancement of intercalative BPDE binding to the synthetic polymer poly(dG-dC)·(dG-dC) (52). Hydrophobic effects (52) or increased molecular polarizability and base stacking (53) derived from the methyl group may facilitate the creation of an intercalation site for BPDE. The increase in BPDE intercalative binding to methylated CpG sites is eventually reflected in the extent of covalent carcinogen–DNA interactions where the B[a]P ring of the (+)-trans adduct is associated with the minor groove. It is possible that formation of the otherwise minor stereoisomers of BPDE adducts is also increased at methylated CpG sequences. Likewise, other DNA adducts that arise through an intercalation mode may form more easily at methylated CpGs. Besides these effects on intercalation, electronic effects provided by the C5 methyl group may be transmitted to the 2-amino nitrogen of the base paired guanine and increase its nucleophilicity (47). Identification of the precise mechanisms underlying this phenomenon is a subject of future studies.
Our data suggest that genomic regions that contain clusters of methylated CpG sites should be preferential targets for BPDE adduct formation. Hypermethylated CpG-rich regions are primarily associated with genes on the Xi chromosome in female cells, and, perhaps, with some imprinted genes. It is possible that the increased BPDE adduct formation in these sequences could have genetic consequences, including impaired replication, enhanced mutagenesis, and a perturbation of the semiconservative copying of the methylation pattern (54, 55), which in turn, could lead to an inappropriate reactivation of the silenced genes.
In conclusion, we have shown that cytosine methylation is a major factor determining the strongly preferential BPDE binding to CpG dinucleotide sites in P53 sequences. It may be speculated that other exogenous and/or endogenous chemical carcinogens react in a similar way. In most cancers, the majority of CpG mutations are C → T transitions commonly ascribed to deamination of 5-mC. However, there is a possibility that other mechanisms are involved. For example, an adduct formed at the guanine 3′ to a 5-mC could direct G → A transition mutations instead of the predominant G → T transversions typical for BPDE. G → A transition mutations would be indistinguishable from C → T transitions on the opposite strand within the CpG sequence. A specific carcinogen that would target methylated CpGs preferentially and cause G → A transition mutations has not been identified as yet, but it is conceivable that such a pathway might be involved in P53 mutagenesis in colon cancer, in which up to 50% of all mutations are transitions at methylated CpG sites.
Acknowledgments
We are grateful to Nicholas Geacintov, Larry Sowers, John Termini, Gerald P. Holmquist, and Timothy O’ Connor for helpful discussions. We thank Steven Bates for help with cell culture work. This study was supported by National Institutes of Health Grants RO1CA65652, PO1CA69449, NSFBIR-9220534, and RO1ES03124.
ABBREVIATIONS
- B[a]P
benzo[a]pyrene
- BPDE
(±) anti-7β,8α-dihydroxy-9α,10α-epoxy-7,8,9,10-tetrahydrobenzo[a]pyrene
- LMPCR
ligation-mediated polymerase chain reaction
- 5-mC
5-methylcytosine
- SAM
S-adenosylmethionine
- Xa
active X chromosome
- Xi
inactive X chromosome
References
- 1.Greenblatt M S, Bennett W P, Hollstein M, Harris C C. Cancer Res. 1994;54:4855–4878. [PubMed] [Google Scholar]
- 2.De Vries E M, Ricke D O, De Vries T N, Hartmann A, Blaszyk H, Liao D, Soussi T, Kovach J S, Sommer S S. Hum Mutat. 1996;7:202–213. doi: 10.1002/(SICI)1098-1004(1996)7:3<202::AID-HUMU4>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 3.Caron de Fromentel C, Soussi T. Genes Chromosomes Cancer. 1992;4:1–15. doi: 10.1002/gcc.2870040102. [DOI] [PubMed] [Google Scholar]
- 4.Levine A J, Wu M C, Chang A, Silver A, Attiyeh E F, Lin J, Epstein C B. Ann NY Acad Sci. 1995;768:111–128. doi: 10.1111/j.1749-6632.1995.tb12115.x. [DOI] [PubMed] [Google Scholar]
- 5.Harris C C. Carcinogenesis. 1996;17:1187–1198. doi: 10.1093/carcin/17.6.1187. [DOI] [PubMed] [Google Scholar]
- 6.Tornaletti S, Pfeifer G P. Oncogene. 1995;10:1593–1499. [PubMed] [Google Scholar]
- 7.Hollstein M, Rice K, Greenblatt M S, Soussi T, Fuchs R, Sorlie T, Hovig E, Smith-Sorensen B, Montesano R, Harris C C. Nucleic Acids Res. 1996;22:3551–3555. [PMC free article] [PubMed] [Google Scholar]
- 8.Denissenko M F, Pao A, Tang M-s, Pfeifer G P. Science. 1996;274:430–432. doi: 10.1126/science.274.5286.430. [DOI] [PubMed] [Google Scholar]
- 9.Hecht S S, Carmella S G, Murphy S E, Foiles P G, Chung F-L. J Cell Biochem Suppl. 1993;17F:27–35. doi: 10.1002/jcb.240531005. [DOI] [PubMed] [Google Scholar]
- 10.Osborne M R, Beland F A, Harvey R G, Brookes P. Int J Cancer. 1976;18:362–368. doi: 10.1002/ijc.2910180315. [DOI] [PubMed] [Google Scholar]
- 11.Jeffrey A M, Jennette K W, Blobstein S H, Weinstein I B, Beland F A, Harvey R G, Kasai H, Miura I, Nakanishi K. J Am Chem Soc. 1976;98:5714–5715. doi: 10.1021/ja00434a060. [DOI] [PubMed] [Google Scholar]
- 12.Meehan T, Straub K. Nature (London) 1979;277:410–412. doi: 10.1038/277410a0. [DOI] [PubMed] [Google Scholar]
- 13.Cheng S C, Hilton B D, Roman J M, Dipple A. Chem Res Toxicol. 1989;2:334–340. doi: 10.1021/tx00011a011. [DOI] [PubMed] [Google Scholar]
- 14.Levin W, Wood A W, Yagi H, Jerina D M, Conney A H. Proc Natl Acad Sci USA. 1976;73:3867–3871. doi: 10.1073/pnas.73.11.3867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Slaga T J, Bracken W J, Gleason G, Levin W, Yagi H, Jerina D M, Conney A H. Cancer Res. 1979;39:67–71. [PubMed] [Google Scholar]
- 16.Riggs A D, Pfeifer G P. Trends Genet. 1992;8:169–174. doi: 10.1016/0168-9525(92)90219-t. [DOI] [PubMed] [Google Scholar]
- 17.Jones P A, Rideout W M, Shen J-C, Spruck C H, Tsai Y C. BioEssays. 1992;14:33–36. doi: 10.1002/bies.950140107. [DOI] [PubMed] [Google Scholar]
- 18.Laird P W, Jaenisch R. Hum Mol Genet. 1994;3:1487–1495. doi: 10.1093/hmg/3.suppl_1.1487. [DOI] [PubMed] [Google Scholar]
- 19.Jones P A. Cancer Res. 1996;56:2463–2467. [PubMed] [Google Scholar]
- 20.Makos M, Nelkin B D, Lerman M I, Latif F, Zbar B, Baylin S B. Proc Natl Acad Sci USA. 1992;89:1929–1933. doi: 10.1073/pnas.89.5.1929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vertino P M, Spillare E A, Harris C C, Baylin S B. Cancer Res. 1993;53:1684–1689. [PubMed] [Google Scholar]
- 22.Merlo A, Herman J G, Mao L, Lee D J, Gabrielson E, Burger P C, Baylin S B, Sidransky D. Nat Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 23.Gama Sosa M A, Slagel V A, Trewyn R W, Oxenhandler R, Kuo K C, Gehrke C W, Ehrlich M. Nucleic Acids Res. 1983;11:6883–6894. doi: 10.1093/nar/11.19.6883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spruck C H, Rideout W M, Jones P A. In: DNA Methylation: Molecular Biology and Biological Significance. Jost J P, Saluz H P, editors. Basel: Birkhäuser; 1993. pp. 487–509. [Google Scholar]
- 25.Rideout W M, Coetzee G A, Olumi A F, Jones P A. Science. 1990;249:1288–1290. doi: 10.1126/science.1697983. [DOI] [PubMed] [Google Scholar]
- 26.Magewu A N, Jones P A. Mol Cell Biol. 1994;14:4225–4232. doi: 10.1128/mcb.14.6.4225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hansen R S, Ellis N A, Gartler S M. Mol Cell Biol. 1988;8:4692–4699. doi: 10.1128/mcb.8.11.4692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pfeifer G P, Singer-Sam J, Riggs A D. Methods Enzymol. 1993;225:567–583. doi: 10.1016/0076-6879(93)25037-3. [DOI] [PubMed] [Google Scholar]
- 29.Lamb P, Crawford L. Mol Cell Biol. 1986;6:1379–1385. doi: 10.1128/mcb.6.5.1379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tang M-s, Pierce J R, Doisy R P, Nazimiec M E, MacLeod M C. Biochemistry. 1992;31:8429–8436. doi: 10.1021/bi00151a006. [DOI] [PubMed] [Google Scholar]
- 31.Venkatachalam S, Denissenko M F, Wani A A. Carcinogenesis. 1995;16:2029–2036. doi: 10.1093/carcin/16.9.2029. [DOI] [PubMed] [Google Scholar]
- 32.Denissenko M F, Venkatachalam S, Ma Y-H, Wani A A. Mutat Res. 1996;363:27–42. doi: 10.1016/0921-8777(95)00059-3. [DOI] [PubMed] [Google Scholar]
- 33.Chen J X, Pao A, Zheng Y, Ye X, Kisleyou A S, Morris R, Slaga T J, Harvey R G, Tang M-s. Biochemistry. 1996;35:9594–9602. doi: 10.1021/bi9604136. [DOI] [PubMed] [Google Scholar]
- 34.Tang M-s. In: Technologies for Detection of DNA Damage and Mutations. Pfeifer G P, editor. New York: Plenum; 1996. pp. 139–153. [Google Scholar]
- 35.Tornaletti S, Rozek D, Pfeifer G P. Oncogene. 1993;8:2051–2057. [PubMed] [Google Scholar]
- 36.Pfeifer G P, Steigerwald S D, Hansen R S, Gartler S M, Riggs A D. Proc Natl Acad Sci USA. 1990;87:8252–8256. doi: 10.1073/pnas.87.21.8252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pfeifer G P, Tanguay R L, Steigerwald S D, Riggs A D. Genes Dev. 1990;4:1277–1287. doi: 10.1101/gad.4.8.1277. [DOI] [PubMed] [Google Scholar]
- 38.Tornaletti S, Pfeifer G P. In: Technologies for Detection of DNA Damage and Mutations. Pfeifer G P, editor. New York: Plenum; 1996. pp. 199–209. [Google Scholar]
- 39.Puisieux A, Lim S, Groopman J, Ozturk M. Cancer Res. 1991;51:6185–6189. [PubMed] [Google Scholar]
- 40.Tornaletti S, Pfeifer G P. Mol Carcinogen. 1996;17:192–201. doi: 10.1002/(SICI)1098-2744(199612)17:4<192::AID-MC2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 41.Smith B L, MacLeod M C. J Biol Chem. 1993;268:20620–20629. [PubMed] [Google Scholar]
- 42.Thrall B D, Mann D B, Smerdon M J, Springer D L. Biochemistry. 1994;33:2210–2216. doi: 10.1021/bi00174a030. [DOI] [PubMed] [Google Scholar]
- 43.Mathison B H, Said B, Shank R C. Carcinogenesis. 1993;14:323–327. doi: 10.1093/carcin/14.2.323. [DOI] [PubMed] [Google Scholar]
- 44.Glickman B W, Schaaper R M, Haseltine W A, Dunn R L, Brash D E. Proc Natl Acad Sci USA. 1986;83:6945–6949. doi: 10.1073/pnas.83.18.6945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pfeifer G P, Drouin R, Riggs A D, Holmquist G P. Proc Natl Acad Sci USA. 1991;88:1374–1378. doi: 10.1073/pnas.88.4.1374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Millard J T, Beachy T M. Biochemistry. 1993;32:12850–12856. doi: 10.1021/bi00210a038. [DOI] [PubMed] [Google Scholar]
- 47.Johnson W S, He Q-Y, Tomasz M. Bioorg Med Chem. 1995;3:851–860. doi: 10.1016/0968-0896(95)00067-q. [DOI] [PubMed] [Google Scholar]
- 48.Smith S S, Kaplan B E, Sowers L C, Newman E M. Proc Natl Acad Sci USA. 1992;89:4744–4748. doi: 10.1073/pnas.89.10.4744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cosman M, de los Santos C, Fiala R, Hingerty B E, Singh S B, Ibanez V, Margulis L A, Live D, Geacintov N E, Broyde S, Patel D J. Proc Natl Acad Sci USA. 1992;89:1914–1918. doi: 10.1073/pnas.89.5.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fountain M A, Krugh T R. Biochemistry. 1995;34:3152–3161. doi: 10.1021/bi00010a004. [DOI] [PubMed] [Google Scholar]
- 51.Geacintov N E. Carcinogenesis. 1986;7:759–766. doi: 10.1093/carcin/7.5.759. [DOI] [PubMed] [Google Scholar]
- 52.Geacintov N E, Shabaz M, Ibanez V, Moussaoui K, Harvey R G. Biochemistry. 1988;27:8380–8387. doi: 10.1021/bi00422a013. [DOI] [PubMed] [Google Scholar]
- 53.Sowers L C, Ramsay Shaw B, Sedwick D. Biochem Biophys Res Commun. 1987;148:790–794. doi: 10.1016/0006-291x(87)90945-4. [DOI] [PubMed] [Google Scholar]
- 54.Wilson V L, Jones P A. Cell. 1983;32:239–246. doi: 10.1016/0092-8674(83)90514-7. [DOI] [PubMed] [Google Scholar]
- 55.Pfeifer G P, Grunberger D, Drahovsky D. Carcinogenesis. 1984;5:931–934. doi: 10.1093/carcin/5.7.931. [DOI] [PubMed] [Google Scholar]