Abstract
Expression of the pyrC gene in Escherichia coli K-12 is regulated by a translational control mechanism in which CTP (and perhaps GTP) pool sizes determine the selection of alternative transcriptional start sites at the pyrC promoter. High CTP levels cause transcription to start primarily at a site that directs the synthesis of untranslatable pyrC transcripts. These transcripts form a hairpin at their 5' ends that blocks ribosome binding to the Shine-Dalgarno (SD) sequence. The pyrC ribosome binding site is unusual in that it contains two potential SD sequences, designated SD1 and SD2, which are located 11 and 4 nucleotides upstream of the translational initiation codon, respectively. In this study, we examined the functions of these two SD sequences in translational initiation. Mutations that inactivate either SD1 or SD2 were constructed and incorporated separately into a pyrC::lacZ protein fusion. The effects of the mutations on pyrC::lacZ expression, regulation, and transcript levels were determined. The results indicate that SD1 is the only functional pyrC SD sequence. The SD2 mutation did cause a small reduction in expression, but this effect appeared to be due to a decrease in transcript stability. In addition, we constructed a mutation that introduces a long spacer region between the hairpin at the 5' end of the pyrC transcript and a new pyrC SD sequence. As predicted by the model for translational control, this mutation caused constitutive expression of a pyrC::lacZ protein fusion.
Full text
PDF

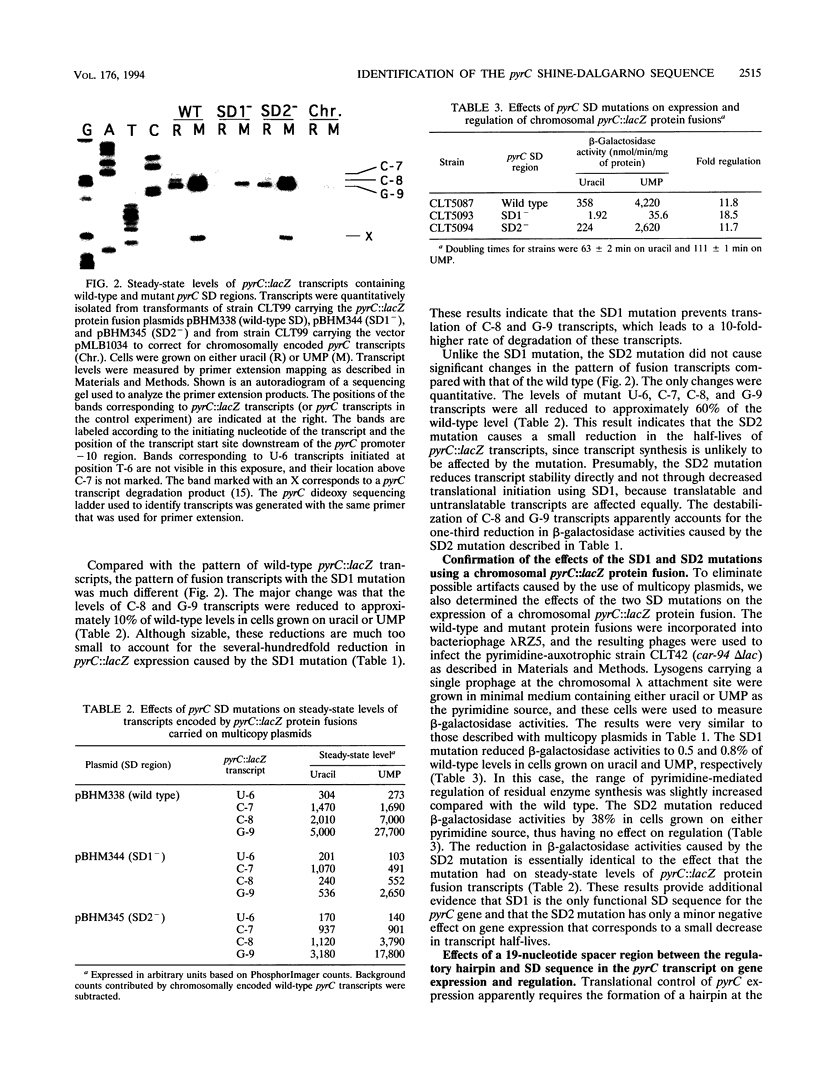

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alper M. D., Ames B. N. Transport of antibiotics and metabolite analogs by systems under cyclic AMP control: positive selection of Salmonella typhimurium cya and crp mutants. J Bacteriol. 1978 Jan;133(1):149–157. doi: 10.1128/jb.133.1.149-157.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frick M. M., Neuhard J., Kelln R. A. Cloning, nucleotide sequence and regulation of the Salmonella typhimurium pyrD gene encoding dihydroorotate dehydrogenase. Eur J Biochem. 1990 Dec 12;194(2):573–578. doi: 10.1111/j.1432-1033.1990.tb15654.x. [DOI] [PubMed] [Google Scholar]
- Gold L. Posttranscriptional regulatory mechanisms in Escherichia coli. Annu Rev Biochem. 1988;57:199–233. doi: 10.1146/annurev.bi.57.070188.001215. [DOI] [PubMed] [Google Scholar]
- Ringquist S., Shinedling S., Barrick D., Green L., Binkley J., Stormo G. D., Gold L. Translation initiation in Escherichia coli: sequences within the ribosome-binding site. Mol Microbiol. 1992 May;6(9):1219–1229. doi: 10.1111/j.1365-2958.1992.tb01561.x. [DOI] [PubMed] [Google Scholar]
- Roland K. L., Liu C. G., Turnbough C. L., Jr Role of the ribosome in suppressing transcriptional termination at the pyrBI attenuator of Escherichia coli K-12. Proc Natl Acad Sci U S A. 1988 Oct;85(19):7149–7153. doi: 10.1073/pnas.85.19.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roland K. L., Powell F. E., Turnbough C. L., Jr Role of translation and attenuation in the control of pyrBI operon expression in Escherichia coli K-12. J Bacteriol. 1985 Sep;163(3):991–999. doi: 10.1128/jb.163.3.991-999.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadler W. C., Switzer R. L. Regulation of Salmonella phosphoribosylpyrophosphate synthetase activity in vivo. Deductions from pool measurements. J Biol Chem. 1977 Dec 10;252(23):8504–8511. [PubMed] [Google Scholar]
- Stormo G. D., Schneider T. D., Gold L. M. Characterization of translational initiation sites in E. coli. Nucleic Acids Res. 1982 May 11;10(9):2971–2996. doi: 10.1093/nar/10.9.2971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen K. I., Baker K. E., Kelln R. A., Neuhard J. Nucleotide pool-sensitive selection of the transcriptional start site in vivo at the Salmonella typhimurium pyrC and pyrD promoters. J Bacteriol. 1993 Jul;175(13):4137–4144. doi: 10.1128/jb.175.13.4137-4144.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen K. I., Neuhard J. Dual transcriptional initiation sites from the pyrC promoter control expression of the gene in Salmonella typhimurium. Mol Gen Genet. 1991 Feb;225(2):249–256. doi: 10.1007/BF00269856. [DOI] [PubMed] [Google Scholar]
- Wilson H. R., Archer C. D., Liu J. K., Turnbough C. L., Jr Translational control of pyrC expression mediated by nucleotide-sensitive selection of transcriptional start sites in Escherichia coli. J Bacteriol. 1992 Jan;174(2):514–524. doi: 10.1128/jb.174.2.514-524.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson H. R., Chan P. T., Turnbough C. L., Jr Nucleotide sequence and expression of the pyrC gene of Escherichia coli K-12. J Bacteriol. 1987 Jul;169(7):3051–3058. doi: 10.1128/jb.169.7.3051-3058.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson H. R., Turnbough C. L., Jr Role of the purine repressor in the regulation of pyrimidine gene expression in Escherichia coli K-12. J Bacteriol. 1990 Jun;172(6):3208–3213. doi: 10.1128/jb.172.6.3208-3213.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Smit M. H., van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990 Oct;87(19):7668–7672. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]



