Abstract
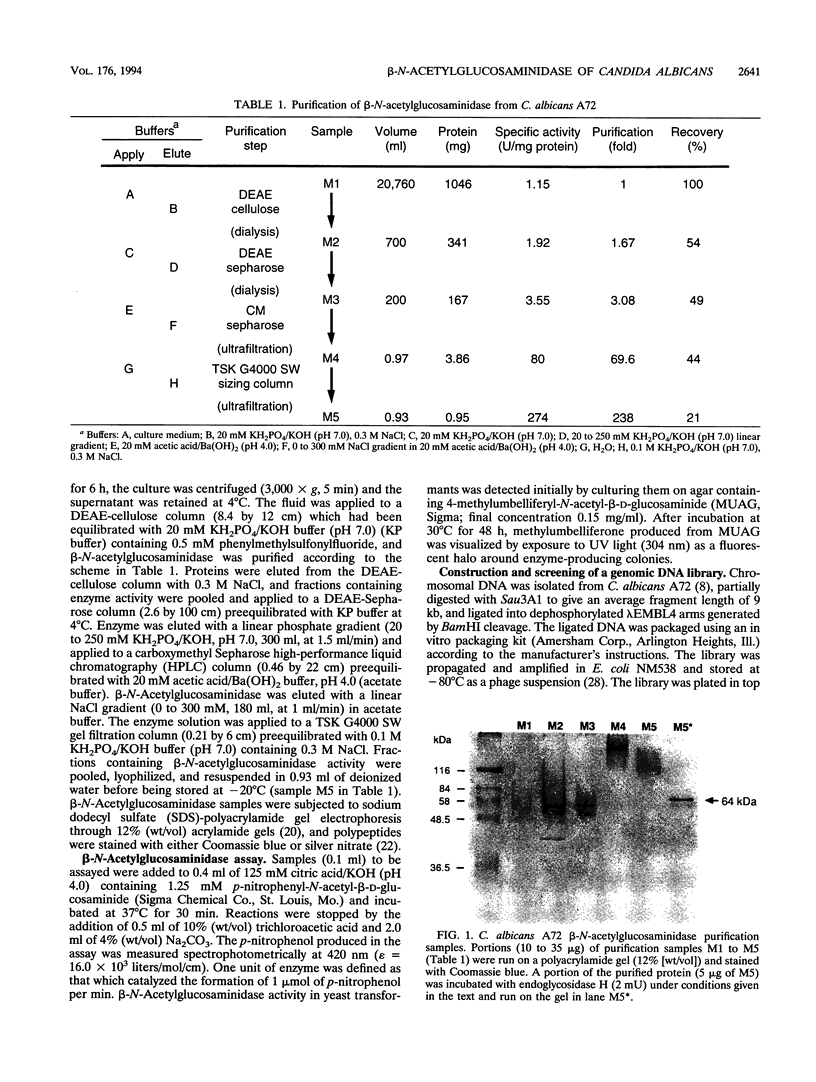
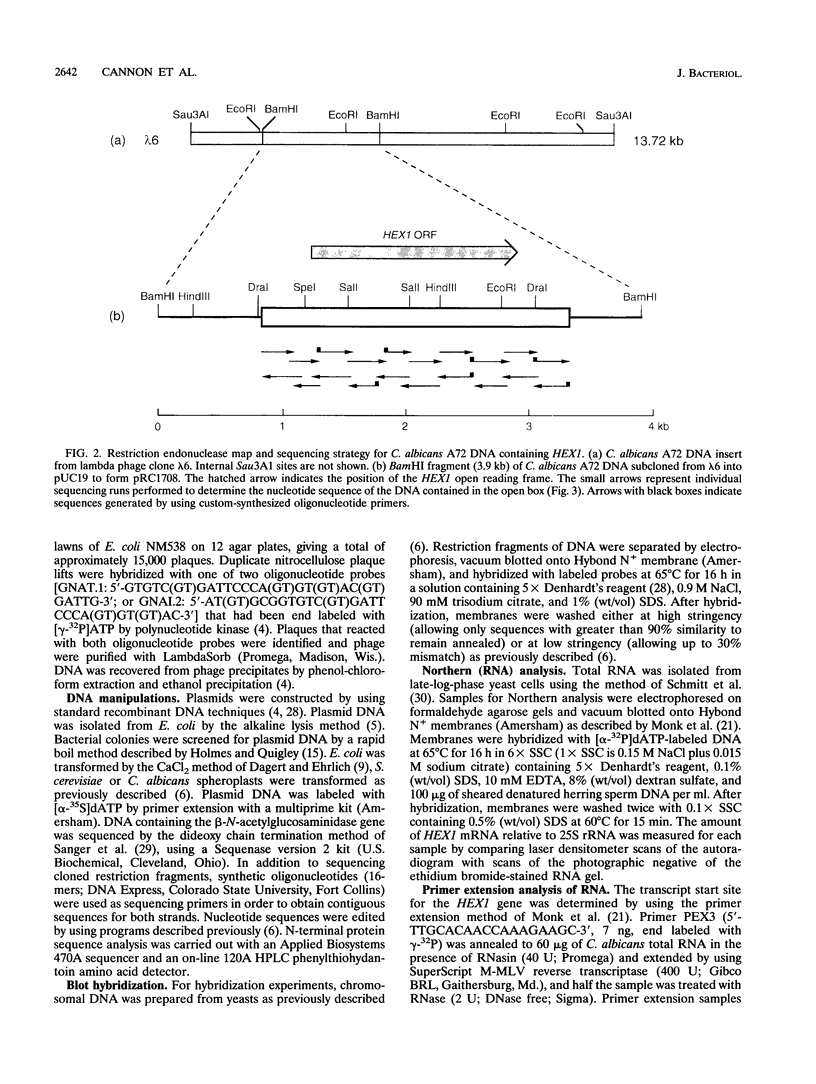
beta-N-Acetylglucosaminidase was purified from the spent culture medium of Candida albicans A72 grown in the presence of N-acetylglucosamine (GlcNAc). The N-terminal amino acid sequence of the protein was determined, two degenerate oligonucleotide probes were constructed, and a 3.9-kb BamHI fragment of DNA that hybridized to both probes was subcloned from a lambda EMBL4 library of C. albicans A72 genomic DNA. This fragment of DNA contained the entire beta-N-acetylglucosaminidase (HEX1) gene, which consisted of an open reading frame coding for a polypeptide precursor of 562 amino acids with a putative 22-amino-acid leader sequence. The deduced HEX1 amino acid sequence showed similarity to hexosaminidases from a variety of organisms. Growth of C. albicans on GlcNAc induced transcription of HEX1, resulting in increased specific beta-N-acetylglucosaminidase activity. HEX1 mRNA (2.35 kb) from GlcNAc-grown cells was approximately 200 bp larger than HEX1 mRNA from cells grown on glucose. This size difference was suggested to result from the use of alternative transcription termination sites. The cloned HEX1 gene introduced into C. albicans SGY-243 on a plasmid also responded to GlcNAc induction.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J. Basic local alignment search tool. J Mol Biol. 1990 Oct 5;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Au-Young J., Robbins P. W. Isolation of a chitin synthase gene (CHS1) from Candida albicans by expression in Saccharomyces cerevisiae. Mol Microbiol. 1990 Feb;4(2):197–207. doi: 10.1111/j.1365-2958.1990.tb00587.x. [DOI] [PubMed] [Google Scholar]
- Bapat B., Ethier M., Neote K., Mahuran D., Gravel R. A. Cloning and sequence analysis of a cDNA encoding the beta-subunit of mouse beta-hexosaminidase. FEBS Lett. 1988 Sep 12;237(1-2):191–195. doi: 10.1016/0014-5793(88)80199-6. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cannon R. D., Jenkinson H. F., Shepherd M. G. Cloning and expression of Candida albicans ADE2 and proteinase genes on a replicative plasmid in C. albicans and in Saccharomyces cerevisiae. Mol Gen Genet. 1992 Nov;235(2-3):453–457. doi: 10.1007/BF00279393. [DOI] [PubMed] [Google Scholar]
- Cannon R. D., Jenkinson H. F., Shepherd M. G. Isolation and nucleotide sequence of an autonomously replicating sequence (ARS) element functional in Candida albicans and Saccharomyces cerevisiae. Mol Gen Genet. 1990 Apr;221(2):210–218. doi: 10.1007/BF00261723. [DOI] [PubMed] [Google Scholar]
- Cryer D. R., Eccleshall R., Marmur J. Isolation of yeast DNA. Methods Cell Biol. 1975;12:39–44. doi: 10.1016/s0091-679x(08)60950-4. [DOI] [PubMed] [Google Scholar]
- Dagert M., Ehrlich S. D. Prolonged incubation in calcium chloride improves the competence of Escherichia coli cells. Gene. 1979 May;6(1):23–28. doi: 10.1016/0378-1119(79)90082-9. [DOI] [PubMed] [Google Scholar]
- Gopal P., Sullivan P. A., Shepherd M. G. Enzymes of N-acetylglucosamine metabolism during germ-tube formation in Candida albicans. J Gen Microbiol. 1982 Oct;128(10):2319–2326. doi: 10.1099/00221287-128-10-2319. [DOI] [PubMed] [Google Scholar]
- Graham T. R., Zassenhaus H. P., Kaplan A. Molecular cloning of the cDNA which encodes beta-N-acetylhexosaminidase A from Dictyostelium discoideum. Complete amino acid sequence and homology with the human enzyme. J Biol Chem. 1988 Nov 15;263(32):16823–16829. [PubMed] [Google Scholar]
- Hahn S., Hoar E. T., Guarente L. Each of three "TATA elements" specifies a subset of the transcription initiation sites at the CYC-1 promoter of Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1985 Dec;82(24):8562–8566. doi: 10.1073/pnas.82.24.8562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann S., Obermaier B., Vogel K., Domdey H. Identification of pre-mRNA polyadenylation sites in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Sep;12(9):4215–4229. doi: 10.1128/mcb.12.9.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes A. R., Cannon R. D., Shepherd M. G. Effect of calcium ion uptake on Candida albicans morphology. FEMS Microbiol Lett. 1991 Jan 15;61(2-3):187–193. doi: 10.1016/0378-1097(91)90549-p. [DOI] [PubMed] [Google Scholar]
- Holmes D. S., Quigley M. A rapid boiling method for the preparation of bacterial plasmids. Anal Biochem. 1981 Jun;114(1):193–197. doi: 10.1016/0003-2697(81)90473-5. [DOI] [PubMed] [Google Scholar]
- Jenkinson H. F., Shepherd M. G. A mutant of Candida albicans deficient in beta-N-acetylglucosaminidase (chitobiase). J Gen Microbiol. 1987 Aug;133(8):2097–2106. doi: 10.1099/00221287-133-8-2097. [DOI] [PubMed] [Google Scholar]
- Kelly R., Miller S. M., Kurtz M. B., Kirsch D. R. Directed mutagenesis in Candida albicans: one-step gene disruption to isolate ura3 mutants. Mol Cell Biol. 1987 Jan;7(1):199–208. doi: 10.1128/mcb.7.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korneluk R. G., Mahuran D. J., Neote K., Klavins M. H., O'Dowd B. F., Tropak M., Willard H. F., Anderson M. J., Lowden J. A., Gravel R. A. Isolation of cDNA clones coding for the alpha-subunit of human beta-hexosaminidase. Extensive homology between the alpha- and beta-subunits and studies on Tay-Sachs disease. J Biol Chem. 1986 Jun 25;261(18):8407–8413. [PubMed] [Google Scholar]
- Laemmli U. K., Favre M. Maturation of the head of bacteriophage T4. I. DNA packaging events. J Mol Biol. 1973 Nov 15;80(4):575–599. doi: 10.1016/0022-2836(73)90198-8. [DOI] [PubMed] [Google Scholar]
- Miller H. Practical aspects of preparing phage and plasmid DNA: growth, maintenance, and storage of bacteria and bacteriophage. Methods Enzymol. 1987;152:145–170. doi: 10.1016/0076-6879(87)52016-x. [DOI] [PubMed] [Google Scholar]
- Monk B. C., Kurtz M. B., Marrinan J. A., Perlin D. S. Cloning and characterization of the plasma membrane H(+)-ATPase from Candida albicans. J Bacteriol. 1991 Nov;173(21):6826–6836. doi: 10.1128/jb.173.21.6826-6836.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J. H. Silver stain for proteins in polyacrylamide gels: a modified procedure with enhanced uniform sensitivity. Anal Biochem. 1981 Nov 1;117(2):307–310. doi: 10.1016/0003-2697(81)90783-1. [DOI] [PubMed] [Google Scholar]
- Myerowitz R., Piekarz R., Neufeld E. F., Shows T. B., Suzuki K. Human beta-hexosaminidase alpha chain: coding sequence and homology with the beta chain. Proc Natl Acad Sci U S A. 1985 Dec;82(23):7830–7834. doi: 10.1073/pnas.82.23.7830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norrander J., Kempe T., Messing J. Construction of improved M13 vectors using oligodeoxynucleotide-directed mutagenesis. Gene. 1983 Dec;26(1):101–106. doi: 10.1016/0378-1119(83)90040-9. [DOI] [PubMed] [Google Scholar]
- Proudfoot N. J., Brownlee G. G. 3' non-coding region sequences in eukaryotic messenger RNA. Nature. 1976 Sep 16;263(5574):211–214. doi: 10.1038/263211a0. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt M. E., Brown T. A., Trumpower B. L. A rapid and simple method for preparation of RNA from Saccharomyces cerevisiae. Nucleic Acids Res. 1990 May 25;18(10):3091–3092. doi: 10.1093/nar/18.10.3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh B., Datta A. Regulation of N-acetylglucosamine uptake in yeast. Biochim Biophys Acta. 1979 Oct 19;557(1):248–258. doi: 10.1016/0005-2736(79)90107-x. [DOI] [PubMed] [Google Scholar]
- Soto-Gil R. W., Zyskind J. W. N,N'-diacetylchitobiase of Vibrio harveyi. Primary structure, processing, and evolutionary relationships. J Biol Chem. 1989 Sep 5;264(25):14778–14783. [PubMed] [Google Scholar]
- Sullivan P. A., McHugh N. J., Romana L. K., Shepherd M. G. The secretion of N-acetylglucosaminidase during germ-tube formation in Candida albicans. J Gen Microbiol. 1984 Sep;130(9):2213–2218. doi: 10.1099/00221287-130-9-2213. [DOI] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]
- von Heijne G. A new method for predicting signal sequence cleavage sites. Nucleic Acids Res. 1986 Jun 11;14(11):4683–4690. doi: 10.1093/nar/14.11.4683. [DOI] [PMC free article] [PubMed] [Google Scholar]