Abstract
Fas ligand (FasL) is a member of the tumor necrosis factor family and induces apoptosis in Fas (CD95)-bearing target cells. In this study, we generated several mAbs that react with mouse FasL (mFasL) and characterized their functional properties. One of these mAbs, K10, specifically reacted with mFasL derived from C57BL/6 (B6) mice, but not that from BALB/c mice as estimated by surface staining and blocking of cytotoxic activities of mFasL transfectants, suggesting a polymorphism of mFasL. Sequence analysis of mFasL cDNA from several strains revealed that BALB/c and DBA/2 mice have three nucleotide differences from the known B6 and C3H sequences, which result in two amino acid substitutions (Thr-184 → Ala-184 and Glu-218 → Gly-218) in the extracellular region. Analysis of the K10 reactivity and genotyping by PCR-restriction fragment length polymorphism revealed that inbred mice segregate into the following two allotypes: mFasL.1 (B6, C3H, MRL, SJL, NOD, NZB, NZW) and mFasL.2 (BALB/c, DBA/1, DBA/2). Interestingly, COS7 cells expressing BALB/c FasL lysed Fas-bearing target cells more efficiently than those expressing B6 FasL. Furthermore, BALB/c-derived CD8-FasL fusion protein, which is composed of the extracellular domains of human CD8α and mFasL, exhibited 9-fold higher specific activity than did B6-derived CD8-FasL. These results suggest that in mFasL.2 mice the Fas/FasL system works more effectively than in mFasL.1 mice.
Fas ligand (FasL), also known as CD95L or APO-1L, is a type II integral membrane protein belonging to a new emerging family of cell surface cytokines including tumor necrosis factor-α (TNF-α), lymphotoxin-α and -β, CD30L, CD40L, CD70 (CD27L), OX40L, 4–1BBL, and TRAIL (APO-2L) (1, 2). Binding of FasL to its receptor Fas (CD95, APO-1) induces apoptotic cell death (1, 2). Fas is expressed in various tissues including the liver, lung, heart, skin, and intestine, whereas FasL is predominantly expressed on activated T and natural killer cells and mediates cytotoxic activity of these cells (1, 3–5). The Fas/FasL system is involved in activation-induced cell death of mature T cells, which suppresses excessive expansion of activated T cells and eliminates autoreactive T cells, and has been implicated in the pathogenesis of autoimmune diseases, hepatitis, graft-versus-host disease, and AIDS (1, 6). Furthermore, FasL is also expressed in the anterior chamber of the eye and on Sertoli cells in the testis, where it contributes to establishing the immune-privileged status of these organs (7, 8).
Recently, we characterized the expression, function, and biochemical nature of human FasL by using several mAbs (9). However, the characterization of mouse FasL (mFasL) has been hampered by lack of specific mAb. In this context, we generated several anti-mFasL mAbs by immunizing hamsters or FasL-deficient gld mice with mFasL transfectants, and estimated the functional properties of mFasL. Unexpectedly, we found an allotypic polymorphism of mFasL recognized by one of these mAbs, which resulted from two amino acid substitutions in the extracellular region in some strains of mice. More importantly, we found that this polymorphism affects the specific activity of mFasL. Physiological and pathological relevances of this finding are discussed.
MATERIALS AND METHODS
Animals.
Four- to 6-week-old female C57BL/6 (B6), C57BL/6 lpr/lpr (B6 lpr), MRL/Mpj (MRL), DBA/1, DBA/2, SJL/j (SJL), NZB, NZW, and NOD mice were obtained from SLC (Shizuoka, Japan). Six-week-old female Armenian hamsters were obtained from Oriental Yeast (Tokyo). Four- to 6-week-old female BALB/c and C3H/He (C3H) mice were obtained from Charles River Breeding Laboratories (Atsugi, Japan). C57BL/6 gld/gld (B6 gld) mice were originally obtained from The Jackson Laboratory and maintained in our animal facilities.
Cells.
Mouse T lymphoma cell lines (L5178Y and WR19L) and human Fas cDNA transfectant (hFas/WR19L) (10) were kindly provided by S. Yonehara (Kyoto University) and cultured in RPMI 1640 medium containing 10% fetal calf serum, 100 μg/ml streptomycin and penicillin, and 2 mM glutamine (culture medium). A mouse B lymphoma A20.2J and L5178Y expressing B6-derived mFasL cDNA (B6 FasL/L5178Y) (11) were also cultured in the culture medium. A CD8+ alloreactive T cell line, POK, derived from perforin (−/−) 129 mice was kindly provided by N. Shinohara (Mitsubishi Kasei institute of Life Science, Tokyo) and maintained as described (12). A keyhole limpet hemocyanin-specific I-Ed-restricted CD4+ T cell clone, BK-1, derived from BALB/c mice was also provided by N. Shinohara and maintained as described (3). An alloreactive CD4+ T cell clone, T16D derived from C3H mice was kindly provided by H. Nariuchi (University of Tokyo) and maintained as described (6).
Anti-CD3-activated splenocytes were prepared by culturing whole splenocytes (3 × 106 cells/ml) in the culture medium on 6-well plates precoated with 10 μg/ml of anti-CD3-mAb (2C11) for 3 days.
Reagents.
Phorbol 12-myristate 13-acetate (PMA) and ionomycin were purchased from Sigma. Anti-mouse CD3 mAb (2C11) was prepared from the hybridoma obtained from the American Type Culture Collection. Anti-human CD8 mAbs, RPA-T8 and biotinylated OKT8, were obtained from PharMingen and Coulter, respectively.
Preparation of mFasL Transfectants.
B6 FasL/L5178Y cells were generated as described (11). B6 FasL cDNA-transfected baby hamster kidney (B6 FasL/BHK) cells were generated in a similar way. Additionally, mFasL cDNAs were prepared by reverse transcription (RT)-PCR from total RNA of anti-CD3-activated C3H, BALB/c, or DBA/2 splenocytes by using an oligonucleotide corresponding to the first six codons as the 5′ primer and that corresponding to the last six codons as the 3′ primer, according to the published sequence (13). The 5′ and 3′ primers were tagged with a XhoI or a NotI site, respectively. After XhoI and NotI digestion, the PCR product of 850 bp was subcloned into pBluescript II SK(+) (Stratagene) and nucleotide sequences were determined by using an Applied Biosystems 373A automated sequencer and fluoresceinated dye terminator cycle sequencing method. The 850-bp cDNAs were then transferred into the XhoI and NotI sites of the pMKITneo vector, kindly provided by K. Maruyama (Tokyo Medical and Dental University). Transient expression of mFasL cDNAs in COS7 cells was performed by using TransIT-LT2 (Pan Vera, Madison, WI) according to the manufacturer’s instructions. L5178Y cells stably expressing BALB/c-derived FasL cDNA were prepared in a similar way as described (11).
Generation of Anti-mFasL mAbs.
Four-week-old female B6 gld mice were immunized by i.p. injection of B6 FasL/L5178Y (1 × 107 cells) three times at 10-day intervals. Three days after final immunization, the splenocytes were fused with P3U1 mouse myeloma cells as described (14). After hypoxanthine/aminopterin/thymidine selection, the antibodies that neutralized the B6 FasL/L5178Y cytotoxicity against hFas/WR19L were screened. One hybridoma producing K10 mAb (mouse IgG2b,κ) was identified by its strong inhibitory effect and cloned by limiting dilution. K10 was purified from ascites by protein G affinity chromatography. Alternatively, Armenian hamsters were immunized with B6 FasL/BHK cells and hamster anti-mFasL mAbs, MFL1∼4, were identified in a similar way.
Flow Cytometry Analysis.
To examine the reactivity of anti-mFasL mAbs against mFasL transfectants, 1 × 106 cells were incubated with 1 μg of mAbs for 1 h at 4°C followed by fluorescein isothiocyanate (FITC)-labeled goat anti-mouse IgG antibody (Caltag, South San Francisco, CA) that crossreacts with hamster IgG. After washing with PBS, the cells were analyzed on a FACScan (Becton Dickinson) and data were processed by using the cellquest program (Becton Dickinson).
3H-TdR Release Assay.
3H-TdR release assay was performed as described (12) with 3H-TdR-labeled target cells (1 × 104) and effector cells at the indicated effector-to-target (E/T) ratios. In the case of bystander cytotoxicity by cytotoxic T lymphocyte clones, 10 ng/ml PMA plus 500 ng/ml ionomycin were added at the start of the cytotoxic assay. After 6 h, intact cells were harvested using Micro 96 Harvester (Skatron, Lier, Norway) and radioactivity was measured on a microplate β counter (Micro Beta Plus, Wallac, Turku, Finland). Percent TdR release was calculated as follows; [(cpm without effector − cpm with effector)/cpm without effector] × 100.
Nucleotide Sequence Analysis.
mFasL cDNAs containing an entire coding region were prepared by RT-PCR from total RNA of anti-CD3-activated splenocytes by using sense and antisense primers corresponding to the 5′ untranslated region (GAGAAGGAAACCCTTTCCTG) and 3′ untranslated region (TGGAAGTGAGTGTAAAGGT) of the published sequence (13). The PCR products were purified by qiaquick-spin (Qiagen, Chatsworth,CA) and both strands were directly sequenced as described above. Three independent PCR products were analyzed to avoid possible artifacts of Taq polymerase.
Comparative Modeling of mFasL.
A three-dimensional model of mFasL monomer was built by modeller (Molecular Simulations, San Diego), a program that implements comparative modeling by satisfaction of spatial restraints. The crystallographic structures of TNF-α and LT-α (Brookhaven Protein Data Bank entries 1TNF and 1TNR) were used as suitable template structures. After modeling, three copies of each monomer were superimposed on the TNF-α trimer (1TNF). The trimer structure was optimized by 100 steps of conjugate gradient minimization using the charmm program (Molecular Simulations).
PCR-Restriction Fragment Length Polymorphism (RFLP).
mFasL cDNAs containing entire coding regions were amplified by RT-PCR from total RNA of anti-CD3-activated splenocytes by using oligonucleotides corresponding to the first six codons as the 5′ primer and to the last six codons as the 3′ primer as described above. After digestion with AluI or Eco81I, PCR products were resolved in 4% NUSIEVE (FMC) gel or 1% agarose gel, respectively.
Construction and Preparation of Soluble CD8-FasL Fusion Proteins.
The cDNA-encoding extracellular region of mFasL (amino acids 137–279) was amplified by PCR from the pBluescript II SK(+) plasmid carrying mFasL cDNA using GAAAAAAAAGAGCCGAGG as the 5′ primer and TTAAAGCTTATACAAGCC as the 3′ primer. EcoRV and NotI sites were introduced into the 5′ and 3′ primers, respectively. After EcoRV and NotI digestion, the PCR product of 430 bp was subcloned into HindIII- and NotI-digested pBluescript II SK(+), together with the 700-bp HindIII–EcoRV fragment encoding the extracellular region of human CD8α, which were derived from pGEM3 carrying human CD8α cDNA (15) (kindly provided by H. Nakauchi, Tsukuba University, Tsukuba). This results in in-frame fusion of the human CD8α extracellular region to mFasL extracellular region (CD8-FasL). After confirmation of the nucleotide sequence, the 1.2-kb XhoI–NotI fragment containing the fusion construct was transferred into XhoI and NotI sites of pMKITneo. COS7 cells were transfected with CD8-FasL/pMKITneo, as described above. At 24 h after transfection, the culture medium was changed to RPMI 1640 containing 1% FCS and further cultured for 96 h. The culture supernatant was collected and concentrated by 10-fold using Vibapore (Vibascience, Binbrook, U.K.). Concentration of CD8-FasL soluble fusion proteins in the supernatant was evaluated by sandwich ELISA using RPA-T8 and biotinylated OKT8. Serially diluted CD8-FasL, which was affinity-purified on OKT8 column, was used as the standard.
Cytotoxic Assays.
Cytotoxic activity of CD8-FasL soluble fusion proteins against hFas/WR19L, WR19L, and A20.2J was tested by the alamar blue method as described (9). Cytotoxic activity of CD8-FasL against thymocytes was tested by the annexin V binding method for apoptotic cells. Thymocytes (2 × 105 cells) were cultured for 24 h with serially diluted CD8-FasL fusion protein in the presence of 10 μg/ml cycloheximide (Sigma). After washing with PBS, the cells were stained with FITC–annexin V (Bender MedSystems, Vienna), according to the manufacturer’s instructions. Apoptotic cells were quantified on FACScan.
RESULTS AND DISCUSSION
Generation of Anti-mFasL mAbs.
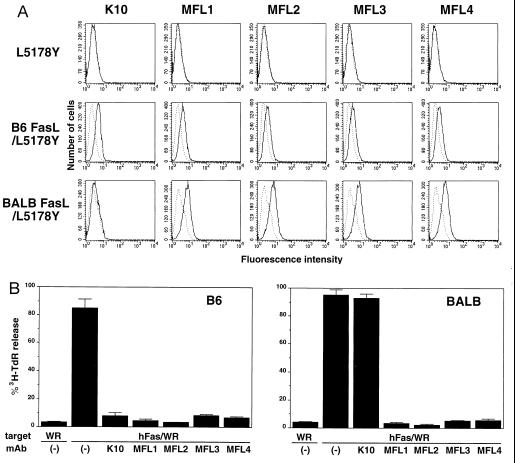
To characterize functional properties of mFasL, we generated several mAbs that specifically bind to mFasL and block its cytotoxic activity. Previous studies have indicated that gld mice have nonfunctional FasL resulting from a single amino acid mutation in the extracellular region (13, 16, 17). We then immunized B6 gld mice with L5178Y cells expressing functional FasL derived from wild-type B6 mice (11). A hybridoma, producing K10 mAb (mouse IgG2b,κ), was selected for its ability to block the B6 FasL/L5178Y cytotoxicity against hFas/WR19L target cells. K10 specifically bound to B6 FasL/L5178Y, but not to L5178Y, as estimated by surface immunofluorescence and flow cytometry (Fig. 1A). We also generated four hamster anti-mFasL mAbs, termed MFL1∼4, that specifically bound to B6 FasL/L5178Y, but not to L5178Y (Fig. 1A). As represented in Fig. 1B, K10 and MFL1∼4 efficiently inhibited the B6 FasL/L5178Y cytotoxicity against hFas/WR19L in a 6-h 3H-TdR release assay.
Figure 1.
Reactivity of anti-mouse FasL mAbs. (A) Cell surface staining of mouse FasL transfectants. L5178Y, B6 FasL/L5178Y, and BALB FasL/L5178Y cells were stained with anti-mouse FasL mAbs, K10, and MFL1∼4, followed by FITC-labeled goat anti-mouse IgG antibody that crossreacts with hamster IgG (solid lines). Broken lines indicate background staining with FITC-labeled goat anti-mouse IgG antibody alone. (B) Blocking of cytotoxic activity of mouse FasL transfectants. Cytotoxic activities of B6 FasL/L5178Y or BALB FasL/L5178Y cells were tested against hFas/WR19L (hFas/WR) or WR19L (WR) target cells in the absence or presence of 10 μg/ml anti-mouse FasL mAbs (K10 and MFL1∼4) in a 6-h 3H-TdR release assay at an E/T = 10. Data represent mean ± SD of triplicate samples.
Inhibition of Fas/FasL-Dependent Cytotoxic Activity of T Cell Clones.
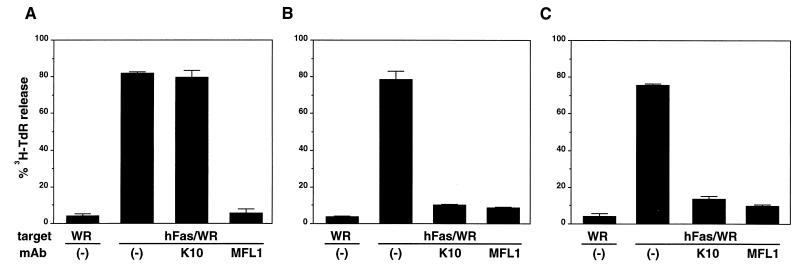
FasL is predominantly expressed on activated T and natural killer cells and mediates Fas-bearing target cell lysis by these effector cells (3–5). It has been demonstrated that some T cell lines exerted Fas/FasL-mediated bystander target cell lysis upon activation with PMA + ionomycin (4, 18, 19). We then tested whether K10 and MFL1∼4 mAbs could block the Fas/FasL-dependent cytotoxic activity of T cell lines. As we reported previously (3, 6, 12), all T cell lines tested here (BK-1 derived from BALB/c, T16D from C3H, and POK from perforin −/− 129) exhibited high cytotoxic activities against Fas+ target cells when activated by PMA + ionomycin (Fig. 2). The cytotoxic activities of both T16D and POK were efficiently blocked by the addition of K10 (Fig. 2 B and C). However, as represented in Fig. 2A, K10 failed to inhibit the cytotoxic activity of BK-1. In contrast, MFL1, as well as MFL2∼4 (not shown), inhibited the cytotoxic activities of all these cell lines (Fig. 2). These results suggested a unique feature of K10 mAb whose reactivity with mFasL may depend on strains of mice.
Figure 2.
Blocking of bystander cytotoxic activity of cytotoxic T lymphocyte clones. Cytotoxic activities of BK-1 (A), T16D (B), or POK (C) cells were tested against WR19L (WR) or hFas/WR19L (hFas/WR) target cells in the presence of PMA + ionomycin and 10 μg/ml anti-mouse FasL mAbs (K10 and MFL1) in a 6-h 3H-TdR release assay at an E/T = 20. Data represent mean ± SD of triplicate samples.
Polymorphism of mFasL.
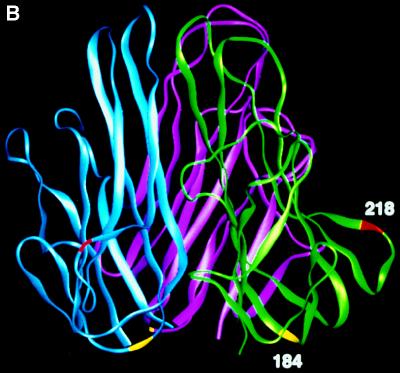
So far, the entire coding region of mFasL cDNA has been determined for 129, C3H, and B6 mice (13, 16, 20) and the amino acid sequences were identical among these strains. To address the possible allotypic difference in mFasL between these strains and BALB/c, we performed direct sequencing of mFasL RT-PCR product from BALB/c mice. BALB/c mice had three nucleotide differences from C3H at nucleotide 550 (A → G), 567 (G → A), and 653 (A → G) (Fig. 3A). The changes at nucleotides 550 and 653 result in Thr to Ala substitution at residue 184 and Glu to Gly substitution at residue 218, respectively, both of which are located in the extracellular region of mFasL. Based on the computer modeling of mFasL structure, Thr-184 and Glu-218 were predicted to locate on the subunit surface at the bottom or the outer tip-forming loop, respectively (Fig. 3B).
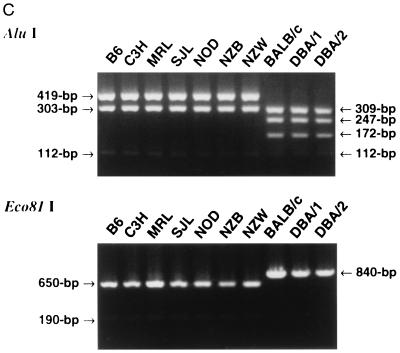
Figure 3.
Polymorphism of mouse FasL. (A) Sequence differences. The upper and lower lines show the nucleotide sequences and predicted amino acid sequences of B6 or BALB/c FasL, respectively. Three stars indicate the nucleotide substitutions found in BALB/c FasL. The changed amino acids (Ala-184 and Gly-218) found in BALB/c FasL are outlined. (B) Side view of mouse FasL trimer. Each subunit is shown in a ribbon diagram in different colors (green, purple, or blue). The changed amino acids (Ala-184 and Gly-218) found in BALB/c FasL are shown in yellow or red, respectively. (C) Genotyping of various mouse strains by PCR-RFLP. The entire coding region of murine FasL was amplified by RT-PCR from total RNA of activated spleen cells from the indicated strains of mice. After digestion with AluI or Eco81I, FasL cDNA was resolved in 4% NUSIEVE gel or 1% agarose gel, respectively.
We next generated a BALB FasL/L5178Y transfectant and tested its reactivity with K10. As represented in the bottom row of Fig. 1A, K10 exhibited little if any binding to BALB FasL/L5178Y as compared with MFL1∼4. Consistently, MFL1∼4, but not K10, blocked the cytotoxic activity of BALB FasL/L5178Y against hFas/WR19L (Fig. 1B). Even at a higher concentration (100 μg/ml), K10 did not inhibit the cytotoxic activity of BALB/c FasL (not shown). These results indicated a polymorphism in mFasL and that K10 preferentially reacts with B6-type FasL, but not or very weakly with BALB/c-type FasL.
The A → G change at nucleotide position 550 and that at nucleotide position 653 result in the appearance of an AluI site (AGCT) and the loss of an Eco81I site (CCTGAGG), respectively. We then examined the polymorphism of mFasL in several inbred strains by PCR-RFLP using AluI or Eco81I. As represented in Fig. 3C, the 112-bp, 303-bp, and 419-bp bands typical for 129, C3H, and B6 were also observed in AluI-digested FasL cDNA from MRL, SJL, NOD, NZB, and NZW mice. On the other hand, the 112-bp, 172-bp, 247-bp, and 309-bp bands typical for BALB/c FasL were also observed in DBA/1 and DBA/2 mice. The Eco81I RFLP concordantly classified these inbred strains into the following two genotypes: mfasl.1 (B6, C3H, MRL, SJL, NOD, NZB, NZW) and mfasl.2 (BALB/c, DBA/1, DBA/2). We also confirmed that DBA/2-derived FasL cDNA does have a nucleic acid sequence identical to that from BALB/c, and that K10 does not bind to DBA/2 FasL transfectants, as estimated by flow cytometry and cytotoxic assay (not shown). In addition, K10 specifically bound to anti-CD3-activated T blasts from mfasl.1 mice, but not to those from mfasl.2 mice (not shown), supporting the propriety of this allotypic classification.
Functional Difference Between mFasL.1 and mFasL.2.
As described above, BALB/c and DBA FasL contain two amino acid differences (Thr-184 → Ala-184 and Glu-218 → Gly-218) from other inbred strains in the extracellular region. Comparative molecular modeling of FasL and other members of the TNF family indicated a high degree of structural similarity between these molecules (20). Previous studies using mutant TNF (21, 22) and crystal structure of TNF receptor/LT-α complex (23) suggested that an outer tip-forming loop, termed a D-E loop, is directly involved in ligand-receptor binding. Based on the structure of human TNF-α (24) and the predicted molecular model of mFasL, Glu-218 was predicted to locate at the D-E loop (Fig. 3B). Considering an alignment of human, mouse, and rat FasL (25), Gly-218 found in mfasl.2 mice corresponds to human Gln-220 and rat Gly-217, respectively. In our recent site-directed mutational analysis of human FasL, a substitution of Gln-220 to Ala greatly reduced the cytotoxic activity (details will be described elsewhere). Therefore, it is likely that amino acid polymorphism at this position may affect the receptor-ligand binding. To determine whether the polymorphism affects the ability of FasL to induce apoptosis in Fas+ target cells, we tested the cytotoxic activities of COS7 cells transfected with either B6- or BALB/c-derived FasL cDNA. In addition to WR19L and hFas/WR19L, mouse B lymphoma A20.2J cells, which highly express Fas and are susceptible to agonistic anti-mFas mAb (3), were used as the target cells. The transient expression levels of B6- and BALB/c-derived FasL on COS7 cells were almost equivalent as estimated by staining with MFL1 (not shown). As represented in Fig. 4, the COS7 cells expressing either B6 or BALB/c FasL exhibited potent cytotoxic activities against hFas/WR19L and A20.2J but not against WR19L in a 6-h 3H-TdR release assay. However, BALB/c FasL cDNA-transfected COS7 cells exhibited significantly higher cytotoxic activity against hFas/WR19L as compared with B6 FasL transfectants. Consistently, the cytotoxic activity of BALB/c FasL cDNA-transfected COS7 cells against A20.2J were significantly higher than that of B6 FasL transfectants. Similar results were obtained in four independent experiments performed in a blind manner. These results suggested that the mFasL.2 molecule has higher cytotoxic activity against human and mouse Fas+ target cells than the mFasL.1 molecule.
Figure 4.
Cytotoxic activities of COS7 cells expressing B6 or BALB/c FasL. B6 or BALB/c FasL cDNA were transiently expressed in COS7 cells. At 72 h after transfection, cytotoxic activities of COS7 cells transfected with B6 (•, ▴, ▪) or BALB/c (○, ▵, □) FasL cDNA were tested against hFas/WR19L (•, ○), A20.2J (▴, ▵), or WR19L (▪, □) target cells in a 6-h 3H-TdR release assay. Data represent mean ± SD of triplicate samples. Similar results were obtained in four independent experiments.
To further compare the specific activities of mFasL.1 and mFasL.2, we constructed soluble fusion proteins composed of the extracellular domain of B6 or BALB/c FasL (amino acids 137–279) and the extracellular domain of human CD8α (CD8-FasL). We previously demonstrated that human FasL is efficiently shed from the cell surface as a functional soluble form (9). In contrast, we could not detect significant cytotoxic activity in the supernatant of mFasL transfectants as also described by others (26). Biochemical basis of this difference is presently unknown. Then we adapted the CD8 fusion strategy, which was successfully used for generating the functional soluble form of CD40L, another member of the TNF family (27). COS7 cells transfected with either B6- or BALB/c-derived CD8-FasL fusion constructs produced comparable amounts of CD8-FasL fusion proteins as estimated by sandwich ELISA using two anti-CD8 mAbs. Immunoprecipitation with anti-CD8 mAb from the supernatants of 35S-Cys/Met-labeled COS7 cells exhibited an expected 40-kDa band under reducing condition and multiple bands of 80-, 120-, and 160-kDa under nonreducing condition, indicating that the CD8-FasL fusion proteins formed oligomers (data not shown). No apparent difference in the oligomer formation was noted between B6- and BALB/c-derived CD8-FasL proteins. We then examined the specific activities of the CD8-FasL fusion proteins by using WR19L, hFas/WR19L, and A20.2J as the target cells. As represented in Fig. 5A, both B6- and BALB/c-derived CD8-FasL fusion proteins exhibited potent cytotoxic activities against hFas/WR19L and A20.2J, but not against WR19L. Notably, BALB/c-derived CD8-FasL exerted about 9-fold higher cytotoxic activity against either hFas/WR19L or A20.2J as compared with B6-derived CD8-FasL.
Figure 5.
Comparison of the cytotoxic activities of B6- and BALB/c-derived CD8-FasL fusion proteins. (A) Cytotoxic activity of soluble CD8-FasL fusion proteins derived from B6 (•, ▴, ▪) or BALB/c (○, ▵, □) was tested against hFas/WR19L (•, ○), A20.2J (▴, ▵), or WR19L (▪, □) cells by the alamar blue method. The abscissa showed the concentration of CD8-FasL fusion proteins, which were determined by sandwich ELISA using anti-CD8 mAbs. Results are expressed as % viability compared with the control wells without CD8-FasL. Data represent mean ± SD of triplicate samples. Similar results were obtained in three independent experiments. (B) Cytotoxic activity of soluble CD8-FasL fusion proteins derived from B6 (•, ▴, ▪) or BALB/c (○, ▵, □) was tested against thymocytes derived from B6 (•, ○), BALB/c (▴, ▵), or B6 lpr (▪, □) mice. Data are indicated as % apoptotic cells determined by the FITC–annexin V staining and represent one of three experiments with similar results.
The results in Figs. 4 and 5 clearly indicate that mFasL.2 has higher specific activity than mFasL.1. This difference may be explained by the following possibilities. As described above, the D-E loop appears to be directly involved in ligand-receptor contact. Therefore, it is possible that Gly-218 → Glu-218 substitution in mFasL may lead to a conformational change of D-E loop or partial loss of receptor contact, which results in reduced binding affinity and cytotoxic activity of mFasL.1. Alternatively, the Thr-184 → Ala-184 substitution may also affect the biological activity of mFasL. The Thr-184 and Ala-184 are predicted to locate on the bottom of the mFasL molecule (Fig. 3B) and seem to have no direct contact with Fas. However, a previous study suggested that the lower half of the TNF molecule plays a crucial role in maintaining the correct conformation of the receptor binding site (22). Therefore, amino acid substitution at this position may indirectly influence the receptor binding. It should be also noted that the Thr-184 → Ala-184 substitution results in the loss of an N-glycosylation site in the mFasL.2 molecule, which may also affect the receptor binding or the trimer formation. These possibilities will be further verified by the point mutational study of mFasL.
Influence of mFas Polymorphism.
It has been reported that mFas receptor is also polymorphic. B6, C3H, and DBA/2 mice (mfas.1) have an His-17 → Arg-17 substitution in the extracellular region of Fas, which is different from MRL and BALB/c mice (mfas.2) (28, 29). Therefore, following four possible combinations of murine Fas and FasL there exists: (i) mFas.1-mFasL.1 in C3H and B6 mice, (ii) mFas.1-mFasL.2 in DBA/2 mice, (iii) mFas.2-mFasL.1 in MRL mice, and (iv) mFas.2-mFasL.2 in BALB/c mice. To examine whether the polymorphism of Fas may also affect the Fas/FasL-induced apoptosis, we tested the cytotoxic activities of B6 and BALB/c CD8-FasL against Fas.1-or Fas.2-bearing thymocytes. It has been shown that most thymocytes highly express Fas and are susceptible to agonistic anti-mouse Fas mAb (30). As represented in Fig. 5B, B6 (mFas.1) and BALB/c (mFas.2) thymocytes, but not B6 lpr thymocytes which do not express Fas (30), were susceptible to either B6 (mFasL.1) or BALB/c (mFasL.2) CD8-FasL in a dose-dependent manner. Particularly, as in the cases of hFas/WR19L and A20.2J, BALB/c CD8-FasL induced apoptosis in both B6- and BALB/c-derived thymocytes about 9-fold more efficiently than B6 CD8-FasL. On the other hand, no apparent difference was observed between B6 and BALB/c thymocytes in their susceptibility to either B6 or BALB/c CD8-FasL. Similar results were also obtained from the experiments using MRL or DBA/2 thymocytes as the target cells (not shown). These results suggest that the polymorphism of mFas does not affect the Fas/FasL interaction. Taken altogether, it seems likely that the Fas/FasL system works more efficiently in mFasL.2 mice than in mFasL.1 mice.
CONCLUSION
In this report, we revealed that the polymorphic mFasL molecules have different biological activities. In mice, a loss of function mutation of FasL in gld mice leads to progressive accumulation of abnormally activated lymphocytes and autoimmune diseases similar to systemic lupus erythematosus (31). Recent studies have indicated that the Fas/FasL system plays a crucial role in the suppression of excessive immune responses and the maintenance of peripheral tolerance (1, 6). It has been well known that non-major histocompatibility complex genetic background of T lymphocytes influences various immune responses, determining either resistance or susceptibility to infection with pathogens and to autoimmune diseases (32, 33). It is interesting to note that FasL is preferentially expressed in T helper 1 cells, which were protective against bacterial infections and pathogenic in some autoimmune diseases, and involved in the activation-induced cell death of this functional subset (6). Therefore, the differential activity of FasL may affect the predominance of T helper 1 in immune responses. In addition, FasL is involved in cytotoxic T lymphocyte-mediated cytotoxicity and may play a role in the pathogenesis of hepatitis and GVHD (1, 3, 4, 6). Therefore, the differential activity of FasL affects the susceptibility to these diseases. It has been also reported that FasL is expressed in the eye and testis where it contributes to the immune-privileged status of these organs (7, 8). Especially in the eye, FasL has been demonstrated to act as an anti-inflammatory in viral infection (7). In contrast, we recently demonstrated that FasL acts as a pro-inflammatory at the skin and in the peritoneal cavity (34, 35). In any such instances, the differential activity of FasL may affect the susceptibility to inflammatory diseases in these organs and tissues. Moreover, it has been recently reported that some tumor cells express FasL, which may contribute to their immune evasion (36, 37), whereas we showed that FasL produced by tumor cells exerted a potent anti-tumor effect (35). In any case, the differential activity of FasL may also affect the susceptibility to tumor development. Taken together, the presently observed mFasL polymorphism that affects biological activity may be a novel genetic factor influencing immune responses and disease susceptibility. It would be interesting to explore the presence of a similar polymorphism in human FasL and disease association.
Acknowledgments
We thank Drs. Shin Yonehara, Hideo Nakauchi, and Nobukata Shinohara for cells and Drs. Yasuyuki Eda and Hirofumi Higuchi for helpful comments and suggestions. This work was supported by grants from the Science and Technology Agency, the Ministry of Education, Science and Culture, and the Ministry of Health, Japan.
ABBREVIATIONS
- FasL
Fas ligand
- mFasL
mouse Fas ligand
- TNF
tumor necrosis factor
- RFLP
restriction fragment length polymorphism
- FITC
fluorescein isothiocyanate
- RT
reverse transcription
- E/T
effector-to-target
References
- 1.Nagata S, Golstein P. Science. 1995;267:1449–1456. doi: 10.1126/science.7533326. [DOI] [PubMed] [Google Scholar]
- 2.Suda T, Takahashi T, Golstein P, Nagata S. Cell. 1993;75:1169–1178. doi: 10.1016/0092-8674(93)90326-l. [DOI] [PubMed] [Google Scholar]
- 3.Hanabuchi S, Koyanagi M, Kawasaki A, Shinohara N, Matsuzawa A, Nishimura Y, Kobayashi Y, Yonehara S, Yagita H, Okumura K. Proc Natl Acad Sci USA. 1994;91:4930–4934. doi: 10.1073/pnas.91.11.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Suda T, Okazaki T, Naito Y, Yokota T, Arai N, Ozaki S, Nakao K, Nagata S. J Immunol. 1995;154:3806–3813. [PubMed] [Google Scholar]
- 5.Arase H, Arase N, Saito T. J Exp Med. 1995;181:1235–1238. doi: 10.1084/jem.181.3.1235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yagita H, Hanabuchi S, Asano Y, Tamura T, Nariuchi H, Okumura K. Immunol Rev. 1995;146:223–239. doi: 10.1111/j.1600-065x.1995.tb00691.x. [DOI] [PubMed] [Google Scholar]
- 7.Griffith T S, Brunner T, Fletcher S M, Green D R, Ferguson T A. Science. 1995;270:1189–1192. doi: 10.1126/science.270.5239.1189. [DOI] [PubMed] [Google Scholar]
- 8.Bellgrau D, Gold D, Selawry H, Moore J, Franzusoff A, Duke R C. Nature (London) 1995;377:630–632. doi: 10.1038/377630a0. [DOI] [PubMed] [Google Scholar]
- 9.Kayagaki N, Kawasaki A, Ebata T, Ohmoto H, Ikeda S, Inoue S, Yoshino K, Okumura K, Yagita H. J Exp Med. 1995;182:1777–1783. doi: 10.1084/jem.182.6.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, Sameshima M, Hase A, Seto Y, Nagata S. Cell. 1991;66:233–243. doi: 10.1016/0092-8674(91)90614-5. [DOI] [PubMed] [Google Scholar]
- 11.Seino K, Kayagaki N, Bashuda H, Okumura K, Yagita H. Int Immunol. 1996;8:1347–1354. doi: 10.1093/intimm/8.9.1347. [DOI] [PubMed] [Google Scholar]
- 12.Kojima H, Shinohara N, Hanaoka S, Someya Shirota Y, Takagaki Y, Ohno H, Saito T, Katayama T, Yagita H, Okumura K, Shinkai Y, Alt F W, Matsuzawa A, Yonehara S, Takayama H. Immunity. 1994;1:357–364. doi: 10.1016/1074-7613(94)90066-3. [DOI] [PubMed] [Google Scholar]
- 13.Takahashi T, Tanaka M, Brannan C I, Jenkins N A, Copeland N G, Suda T, Nagata S. Cell. 1994;76:969–976. doi: 10.1016/0092-8674(94)90375-1. [DOI] [PubMed] [Google Scholar]
- 14.Kato K, Koyanagi M, Okada H, Takahashi T, Wong Y W, Williams A F, Okumura K, Yagita H. J Exp Med. 1992;176:1241–1249. doi: 10.1084/jem.176.5.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Giblin P, Ledbetter J A, Kavathas P. Proc Natl Acad Sci USA. 1989;86:998–1002. doi: 10.1073/pnas.86.3.998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lynch D H, Watson M L, Alderson M R, Baum P R, Miller R E, Tough T, Gibson M, Davis Smith T, Smith C A, Hunter K, Bhat D, Din W, Goodwin R G, Seldin M. Immunity. 1994;1:131–136. doi: 10.1016/1074-7613(94)90106-6. [DOI] [PubMed] [Google Scholar]
- 17.Hahne M, Peitsch M C, Irmler M, Schröter M, Lowin B, Rousseau M, Bron C, Renno T, French L, Tschopp J. Int Immunol. 1995;7:1381–1386. doi: 10.1093/intimm/7.9.1381. [DOI] [PubMed] [Google Scholar]
- 18.Rouvier E, Luciani M F, Golstein P. J Exp Med. 1993;177:195–200. doi: 10.1084/jem.177.1.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Vignaux F, Golstein P. Eur J Immunol. 1994;24:923–927. doi: 10.1002/eji.1830240421. [DOI] [PubMed] [Google Scholar]
- 20.Peitsch M C, Tschopp J. Mol Immunol. 1995;32:761–772. doi: 10.1016/0161-5890(95)00016-8. [DOI] [PubMed] [Google Scholar]
- 21.Goh C R, Loh C S, Porter A G. Protein Eng. 1991;4:785–791. doi: 10.1093/protein/4.7.785. [DOI] [PubMed] [Google Scholar]
- 22.van Ostade X, Tavernier J, Prange T, Fiers W. EMBO J. 1991;10:827–836. doi: 10.1002/j.1460-2075.1991.tb08015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Banner D W, D’Arcy A, Janes W, Gentz R, Schoenfeld H, Broger C, Loetscher H, Lesslauer W. Cell. 1993;73:431–445. doi: 10.1016/0092-8674(93)90132-a. [DOI] [PubMed] [Google Scholar]
- 24.Jones E Y, Stuart D I, Walker N P C. Nature (London) 1989;338:225–228. doi: 10.1038/338225a0. [DOI] [PubMed] [Google Scholar]
- 25.Takahashi T, Tanaka M, Inazawa J, Abe T, Suda T, Nagata S. Int Immunol. 1994;6:1567–1574. doi: 10.1093/intimm/6.10.1567. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka M, Suda T, Takahashi T, Nagata S. EMBO J. 1995;14:1129–1135. doi: 10.1002/j.1460-2075.1995.tb07096.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lane P, Brocker T, Hubele S, Padovan E, Lanzavecchia A, McConnell F. J Exp Med. 1993;177:1209–1213. doi: 10.1084/jem.177.4.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drappa J, Brot N, Elkon K B. Proc Natl Acad Sci USA. 1993;90:10340–10344. doi: 10.1073/pnas.90.21.10340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nishimura Y, Ishii A, Kobayashi Y, Yamasaki Y, Yonehara S. J Immunol. 1995;154:4395–4403. [PubMed] [Google Scholar]
- 30.Ogasawara J, Suda T, Nagata S. J Exp Med. 1995;181:485–491. doi: 10.1084/jem.181.2.485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cohen P L, Eisenberg R A. Annu Rev Immunol. 1991;9:243–269. doi: 10.1146/annurev.iy.09.040191.001331. [DOI] [PubMed] [Google Scholar]
- 32.Mosmann T R, Sad S. Immunol Today. 1996;17:138–146. doi: 10.1016/0167-5699(96)80606-2. [DOI] [PubMed] [Google Scholar]
- 33.Liblau R S, Singer S M, McDevitt H O. Immunol Today. 1995;16:34–38. doi: 10.1016/0167-5699(95)80068-9. [DOI] [PubMed] [Google Scholar]
- 34.Yagita H, Seino K, Kayagaki N, Okumura K. Nature (London) 1996;379:682. doi: 10.1038/379682a0. [DOI] [PubMed] [Google Scholar]
- 35.Seino K, Kayagaki N, Okumura K, Yagita H. Nat Med. 1997;3:165–170. doi: 10.1038/nm0297-165. [DOI] [PubMed] [Google Scholar]
- 36.Hahne M, Rimoldi D, Schröter M, Romero P, Schreier M, French L E, Schneider P, Bornand T, Fontana A, Lienard D, Cerottini J-C, Tschopp J. Science. 1996;274:1363–1366. doi: 10.1126/science.274.5291.1363. [DOI] [PubMed] [Google Scholar]
- 37.Strand S, Hofmann W, Hug H, Müller M, Otto G, Strand D, Mariani S M, Stremmel W, Krammer P H, Galle P R. Nat Med. 1996;2:1361–1366. doi: 10.1038/nm1296-1361. [DOI] [PubMed] [Google Scholar]