Abstract
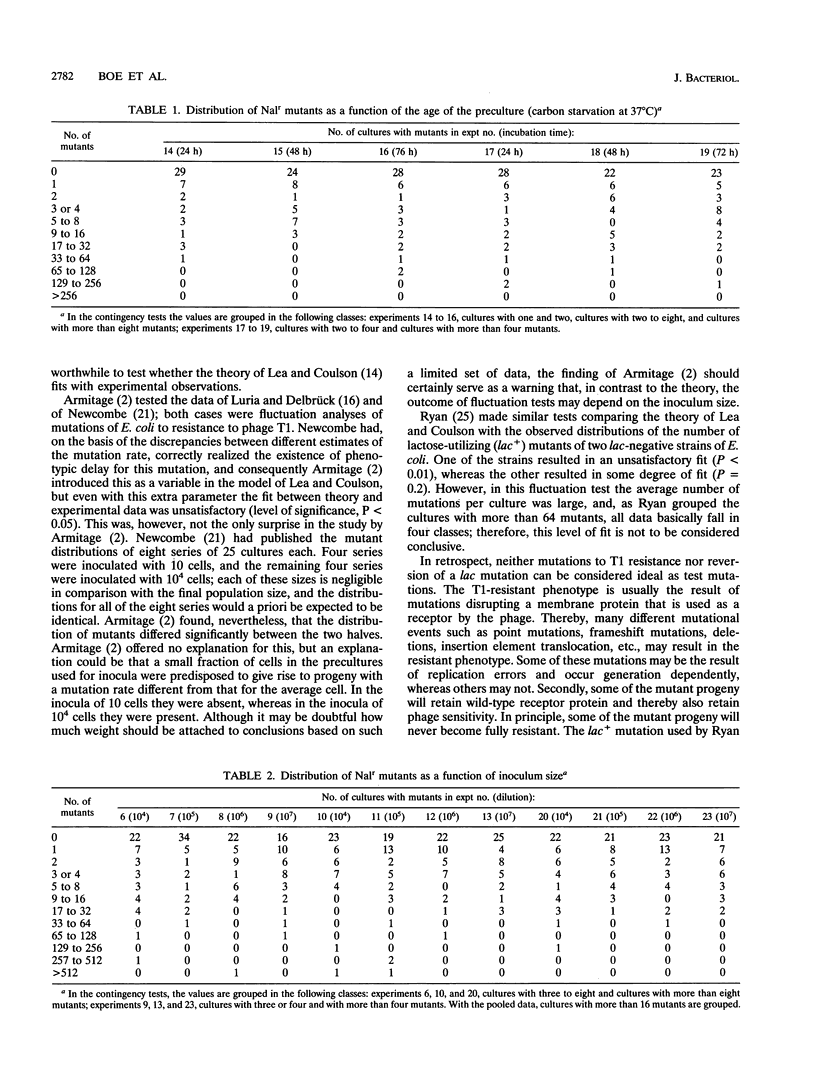
Mutations of Escherichia coli from sensitivity to nalidixic acid resistance were studied by fluctuation analysis. The mutant distributions in replicate cultures were not significantly affected either by the age of the carbon-starved preculture used for inocula or by the inoculum size. The data from 23 fluctuation tests (48 cultures each) were pooled. The mean number of mutations per culture was estimated to be 0.71 from the fraction of cultures without mutants or 0.74 and 0.77 by maximum-likelihood estimation based on the two models under consideration. When the pooled data were compared with the theoretical expectations, the fits were unsatisfactory (P < 0.005). The lack of fit was caused mainly by too high a frequency of cultures with between 17 and 32 mutants and too high a frequency of cultures with more than 128 mutants. Possible reasons for the lack of fit and its implications with respect to estimation of mutation rates from fluctuation tests are discussed.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ARMITAGE P. Statistical concepts in the theory of bacterial mutation. J Hyg (Lond) 1953 Jun;51(2):162–184. doi: 10.1017/s0022172400015606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson R. P., Roth J. R. Tandem genetic duplications in phage and bacteria. Annu Rev Microbiol. 1977;31:473–505. doi: 10.1146/annurev.mi.31.100177.002353. [DOI] [PubMed] [Google Scholar]
- BERTANI G. Studies on lysogenesis. I. The mode of phage liberation by lysogenic Escherichia coli. J Bacteriol. 1951 Sep;62(3):293–300. doi: 10.1128/jb.62.3.293-300.1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boe L. Translational errors as the cause of mutations in Escherichia coli. Mol Gen Genet. 1992 Feb;231(3):469–471. doi: 10.1007/BF00292717. [DOI] [PubMed] [Google Scholar]
- Cairns J., Overbaugh J., Miller S. The origin of mutants. Nature. 1988 Sep 8;335(6186):142–145. doi: 10.1038/335142a0. [DOI] [PubMed] [Google Scholar]
- Cavalli-Sforza L L, Lederberg J. Isolation of Pre-Adaptive Mutants in Bacteria by Sib Selection. Genetics. 1956 May;41(3):367–381. doi: 10.1093/genetics/41.3.367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu J., Li I. C., Chu E. H. The parameters for quantitative analysis of mutation rates with cultured mammalian somatic cells. Mutat Res. 1982 Nov;105(5):363–370. doi: 10.1016/0165-7992(82)90108-7. [DOI] [PubMed] [Google Scholar]
- Kendal W. S., Frost P. Pitfalls and practice of Luria-Delbrück fluctuation analysis: a review. Cancer Res. 1988 Mar 1;48(5):1060–1065. [PubMed] [Google Scholar]
- Li I. C., Wu S. C., Fu J., Chu E. H. A deterministic approach for the estimation of mutation rates in cultured mammalian cells. Mutat Res. 1985 Mar;149(1):127–132. doi: 10.1016/0027-5107(85)90017-x. [DOI] [PubMed] [Google Scholar]
- Luria S. E., Delbrück M. Mutations of Bacteria from Virus Sensitivity to Virus Resistance. Genetics. 1943 Nov;28(6):491–511. doi: 10.1093/genetics/28.6.491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R., Gellert M. Regulation of the genes for E. coli DNA gyrase: homeostatic control of DNA supercoiling. Cell. 1983 Aug;34(1):105–113. doi: 10.1016/0092-8674(83)90140-x. [DOI] [PubMed] [Google Scholar]
- Newcombe H. B. Delayed Phenotypic Expression of Spontaneous Mutations in Escherichia Coli. Genetics. 1948 Sep;33(5):447–476. doi: 10.1093/genetics/33.5.447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ninio J. Transient mutators: a semiquantitative analysis of the influence of translation and transcription errors on mutation rates. Genetics. 1991 Nov;129(3):957–962. doi: 10.1093/genetics/129.3.957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- POWELL E. O. An outline of the pattern of bacterial generation times. J Gen Microbiol. 1958 Apr;18(2):382–417. doi: 10.1099/00221287-18-2-382. [DOI] [PubMed] [Google Scholar]
- POWELL E. O. Growth rate and generation time of bacteria, with special reference to continuous culture. J Gen Microbiol. 1956 Dec;15(3):492–511. doi: 10.1099/00221287-15-3-492. [DOI] [PubMed] [Google Scholar]
- RAYN E. J. Distribution of numbers of mutant bacteria in replicate cultures. Nature. 1952 May 24;169(4308):882–883. doi: 10.1038/169882b0. [DOI] [PubMed] [Google Scholar]
- Sarkar S. Haldane's solution of the Luria-Delbrück distribution. Genetics. 1991 Feb;127(2):257–261. doi: 10.1093/genetics/127.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Ma W. T., Sandri G. H. On fluctuation analysis: a new, simple and efficient method for computing the expected number of mutants. Genetica. 1992;85(2):173–179. doi: 10.1007/BF00120324. [DOI] [PubMed] [Google Scholar]
- Stewart F. M., Gordon D. M., Levin B. R. Fluctuation analysis: the probability distribution of the number of mutants under different conditions. Genetics. 1990 Jan;124(1):175–185. doi: 10.1093/genetics/124.1.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi J., Yoshida H., Yamayoshi M., Nakamura S. Nalidixic acid-resistant mutations of the gyrB gene of Escherichia coli. Mol Gen Genet. 1986 Sep;204(3):367–373. doi: 10.1007/BF00331012. [DOI] [PubMed] [Google Scholar]
- Yoshida H., Kojima T., Yamagishi J., Nakamura S. Quinolone-resistant mutations of the gyrA gene of Escherichia coli. Mol Gen Genet. 1988 Jan;211(1):1–7. doi: 10.1007/BF00338386. [DOI] [PubMed] [Google Scholar]


