Abstract
Aromatic amine dehydrogenase (AADH) catalyzes the oxidative deamination of aromatic amines including tyramine and dopamine. AADH is structurally similar to methylamine dehydrogenase (MADH) and possesses the same tryptophan tryptophylquinone (TTQ) prosthetic group. AADH exhibits an alpha 2 beta 2 structure with subunit molecular weights of 39,000 and 18,000 and with a quinone covalently attached to each beta subunit. Neither subunit cross-reacted immunologically with antibodies to the corresponding subunits of MADH, and the N-terminal amino acid sequence of the beta subunit of AADH exhibited no homology with the highly conserved beta subunits of MADH. The absorption spectra for the oxidized, semiquinone, and reduced forms of AADH have been characterized, and extinction coefficients for the absorption maxima of each redox form have been determined. These spectra are very similar to those for MADH, indicating the likelihood of a TTQ cofactor. This was verified by the near identity of the vibrational frequencies and intensities in the resonance Raman spectra for the oxidized forms of AADH and MADH. A stable semiquinone of AADH could be observed during a reductive titration with dithionite, whereas titration with tyramine proceeded directly from the oxidized to the reduced form. AADH was very stable against denaturation by heat and exposure to guanidine. The individual subunits could be separated by gel filtration after incubation in guanidine hydrochloride, and partial reconstitution of activity was observed on recombination of the subunits. Steady-state kinetic analysis of AADH yielded a Vmax of 17 mumol/min/mg and a Km for tyramine of 5.4 microM. Substrate inhibition by tyramine was observed. AADH was irreversibly inhibited by hydrazine, phenylhydrazine, hydroxylamine, semicarbazide, and aminoguanidine. Isonicotinic acid hydrazide (isoniazid) and isonicotinic acid 2-isopropyl hydrazide (iproniazid) were reversible noncompetitive inhibitors of AADH and exhibited K(i) values of 8 and 186 microM, respectively. The similarities and differences between AADH and other amine oxidizing enzymes are also discussed.
Full text
PDF



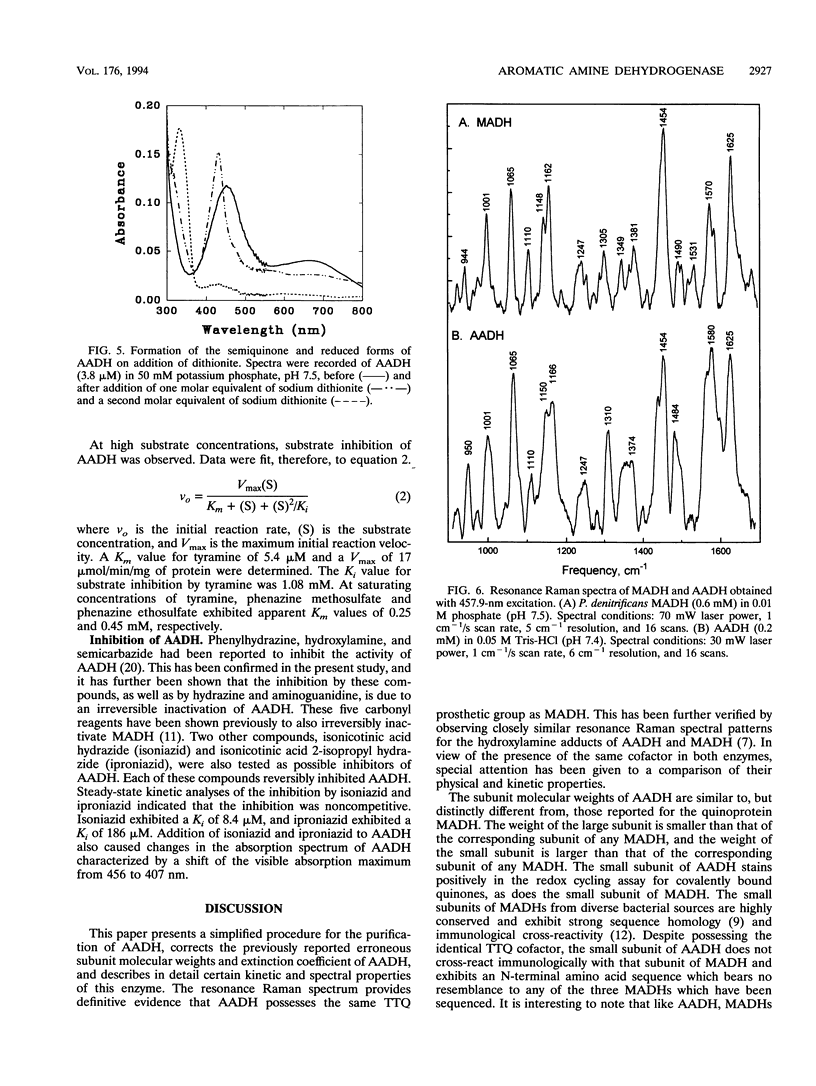


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Backes G., Davidson V. L., Huitema F., Duine J. A., Sanders-Loehr J. Characterization of the tryptophan-derived quinone cofactor of methylamine dehydrogenase by resonance Raman spectroscopy. Biochemistry. 1991 Sep 24;30(38):9201–9210. doi: 10.1021/bi00102a011. [DOI] [PubMed] [Google Scholar]
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Chen L. Y., Mathews F. S., Davidson V. L., Huizinga E. G., Vellieux F. M., Duine J. A., Hol W. G. Crystallographic investigations of the tryptophan-derived cofactor in the quinoprotein methylamine dehydrogenase. FEBS Lett. 1991 Aug 5;287(1-2):163–166. doi: 10.1016/0014-5793(91)80041-z. [DOI] [PubMed] [Google Scholar]
- Chen L., Mathews F. S., Davidson V. L., Huizinga E. G., Vellieux F. M., Hol W. G. Three-dimensional structure of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans determined by molecular replacement at 2.8 A resolution. Proteins. 1992 Oct;14(2):288–299. doi: 10.1002/prot.340140214. [DOI] [PubMed] [Google Scholar]
- Chistoserdov A. Y., Tsygankov Y. D., Lidstrom M. E. Cloning and sequencing of the structural gene for the small subunit of methylamine dehydrogenase from Methylobacterium extorquens AM1: evidence for two tryptophan residues involved in the active center. Biochem Biophys Res Commun. 1990 Oct 15;172(1):211–216. doi: 10.1016/s0006-291x(05)80195-0. [DOI] [PubMed] [Google Scholar]
- Chistoserdov A. Y., Tsygankov Y. D., Lidstrom M. E. Genetic organization of methylamine utilization genes from Methylobacterium extorquens AM1. J Bacteriol. 1991 Sep;173(18):5901–5908. doi: 10.1128/jb.173.18.5901-5908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davidson V. L., Jones L. H. Cofactor-directed inactivation by nucleophilic amines of the quinoprotein methylamine dehydrogenase from Paracoccus denitrificans. Biochim Biophys Acta. 1992 May 22;1121(1-2):104–110. doi: 10.1016/0167-4838(92)90343-c. [DOI] [PubMed] [Google Scholar]
- Davidson V. L. Methylamine dehydrogenases from methylotrophic bacteria. Methods Enzymol. 1990;188:241–246. doi: 10.1016/0076-6879(90)88040-h. [DOI] [PubMed] [Google Scholar]
- Eady R. R., Large P. J. Microbial oxidation of amines. Spectral and kinetic properties of the primary amine dehydrogenase of Pseudomonas AM1. Biochem J. 1971 Aug;123(5):757–771. doi: 10.1042/bj1230757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eady R. R., Large P. J. Purification and properties of an amine dehydrogenase from Pseudomonas AM1 and its role in growth on methylamine. Biochem J. 1968 Jan;106(1):245–255. doi: 10.1042/bj1060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain M., Davidson V. L., Gray K. A., Knaff D. B. Redox properties of the quinoprotein methylamine dehydrogenase from paracoccus denitrificans. Biochemistry. 1987 Jun 30;26(13):4139–4143. doi: 10.1021/bi00387a059. [DOI] [PubMed] [Google Scholar]
- Husain M., Davidson V. L. Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans. J Bacteriol. 1987 Apr;169(4):1712–1717. doi: 10.1128/jb.169.4.1712-1717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishii Y., Hase T., Fukumori Y., Matsubara H., Tobari J. Amino acid sequence studies of the light subunit of methylamine dehydrogenase from Pseudomonas AM1: existence of two residues binding the prosthetic group. J Biochem. 1983 Jan;93(1):107–119. doi: 10.1093/oxfordjournals.jbchem.a134144. [DOI] [PubMed] [Google Scholar]
- Iwaki M., Yagi T., Horiike K., Saeki Y., Ushijima T., Nozaki M. Crystallization and properties of aromatic amine dehydrogenase from Pseudomonas sp. Arch Biochem Biophys. 1983 Jan;220(1):253–262. doi: 10.1016/0003-9861(83)90408-3. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- MARMUR J., DOTY P. Determination of the base composition of deoxyribonucleic acid from its thermal denaturation temperature. J Mol Biol. 1962 Jul;5:109–118. doi: 10.1016/s0022-2836(62)80066-7. [DOI] [PubMed] [Google Scholar]
- McIntire W. S., Bates J. L., Brown D. E., Dooley D. M. Resonance Raman spectroscopy of methylamine dehydrogenase from bacterium W3A1. Biochemistry. 1991 Jan 8;30(1):125–133. doi: 10.1021/bi00215a019. [DOI] [PubMed] [Google Scholar]
- McIntire W. S., Wemmer D. E., Chistoserdov A., Lidstrom M. E. A new cofactor in a prokaryotic enzyme: tryptophan tryptophylquinone as the redox prosthetic group in methylamine dehydrogenase. Science. 1991 May 10;252(5007):817–824. doi: 10.1126/science.2028257. [DOI] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Moos M., Jr, Nguyen N. Y., Liu T. Y. Reproducible high yield sequencing of proteins electrophoretically separated and transferred to an inert support. J Biol Chem. 1988 May 5;263(13):6005–6008. [PubMed] [Google Scholar]
- Mozdzanowski J., Hembach P., Speicher D. W. High yield electroblotting onto polyvinylidene difluoride membranes from polyacrylamide gels. Electrophoresis. 1992 Jan-Feb;13(1-2):59–64. doi: 10.1002/elps.1150130112. [DOI] [PubMed] [Google Scholar]
- Murooka Y., Doi N., Harada T. Distribution of membrane-bound monoamine oxidase in bacteria. Appl Environ Microbiol. 1979 Oct;38(4):565–569. doi: 10.1128/aem.38.4.565-569.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols R., Weaver C. D., Eisenstein E., Blakley R. L., Appleman J., Huang T. H., Huang F. Y., Howell E. E. Titration of histidine 62 in R67 dihydrofolate reductase is linked to a tetramer<-->two-dimer equilibrium. Biochemistry. 1993 Feb 23;32(7):1695–1706. doi: 10.1021/bi00058a002. [DOI] [PubMed] [Google Scholar]
- Nozaki M. Aromatic amine dehydrogenase from Alcaligenes faecalis. Methods Enzymol. 1987;142:650–655. doi: 10.1016/s0076-6879(87)42077-6. [DOI] [PubMed] [Google Scholar]
- Patek D. R., Hellerman L. Mitochondrial monoamine oxidase. Mechanism of inhibition by phenylhydrazine and by aralkylhydrazines. Role of enzymatic oxidation. J Biol Chem. 1974 Apr 25;249(8):2373–2380. [PubMed] [Google Scholar]
- Paz M. A., Flückiger R., Boak A., Kagan H. M., Gallop P. M. Specific detection of quinoproteins by redox-cycling staining. J Biol Chem. 1991 Jan 15;266(2):689–692. [PubMed] [Google Scholar]
- Roark D. E. Sedimentation equilibrium techniques: multiple speed analyses and an overspeed procedure. Biophys Chem. 1976 Jul;5(1-2):185–196. doi: 10.1016/0301-4622(76)80034-8. [DOI] [PubMed] [Google Scholar]
- SMITH T. E., WEISSBACH H., UDENFRIEND S. STUDIES ON MONOAMINE OXIDASE: THE MECHANISM OF INHIBITION OF MONOAMINE OXIDASE BY IPRONIAZID. Biochemistry. 1963 Jul-Aug;2:746–751. doi: 10.1021/bi00904a021. [DOI] [PubMed] [Google Scholar]
- Shirai S., Matsumoto T., Tobari J. Methylamine dehydrogenase of Pseudomonas AM1. A subunit enzyme. J Biochem. 1978 Jun;83(6):1599–1607. doi: 10.1093/oxfordjournals.jbchem.a132071. [DOI] [PubMed] [Google Scholar]
- Van Beeumen J., Van Driessche G., Huitema F., Duine J. A., Canters G. W. N-terminal heterogeneity of methylamine dehydrogenase from Thiobacillus versutus. FEBS Lett. 1993 Oct 25;333(1-2):188–192. doi: 10.1016/0014-5793(93)80402-g. [DOI] [PubMed] [Google Scholar]
- Vellieux F. M., Huitema F., Groendijk H., Kalk K. H., Jzn J. F., Jongejan J. A., Duine J. A., Petratos K., Drenth J., Hol W. G. Structure of quinoprotein methylamine dehydrogenase at 2.25 A resolution. EMBO J. 1989 Aug;8(8):2171–2178. doi: 10.1002/j.1460-2075.1989.tb08339.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Beer R., Duine J. A., Frank J., Large P. J. The prosthetic group of methylamine dehydrogenase from Pseudomonas AM1: evidence for a quinone structure. Biochim Biophys Acta. 1980 Apr 25;622(2):370–374. doi: 10.1016/0005-2795(80)90050-1. [DOI] [PubMed] [Google Scholar]