Abstract
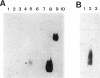
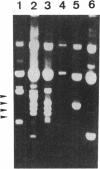
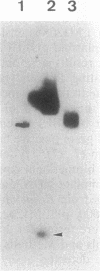
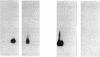
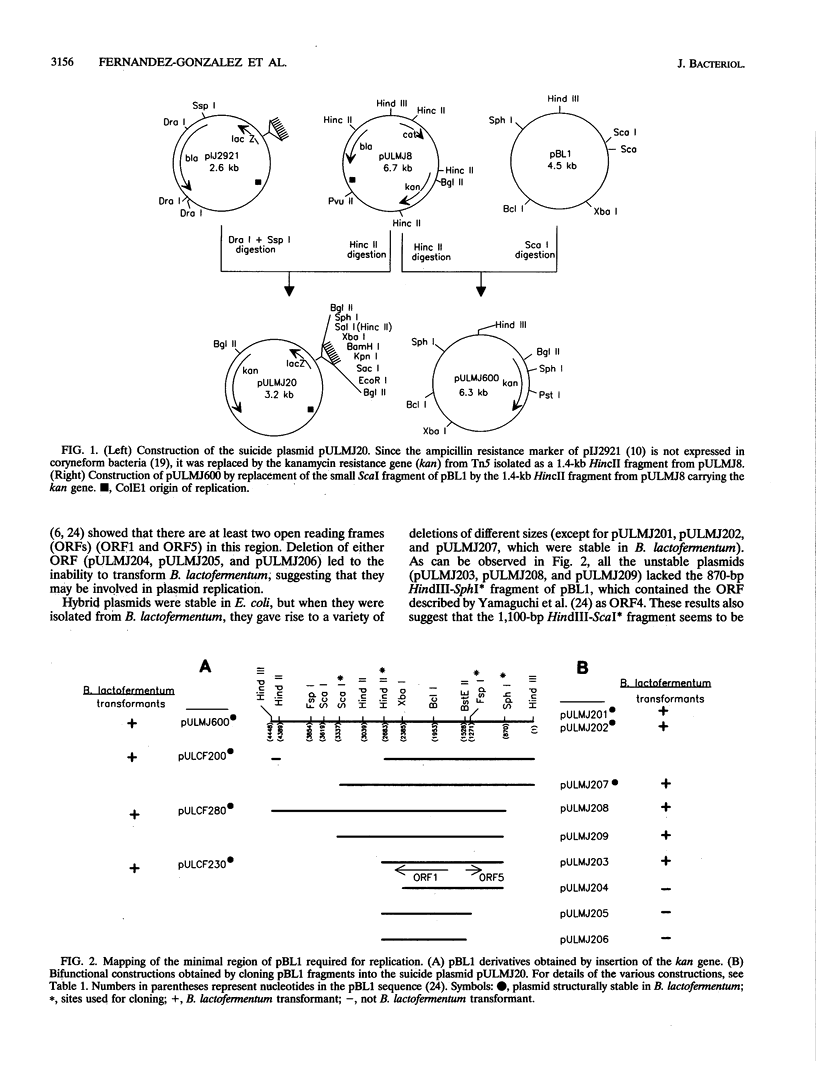
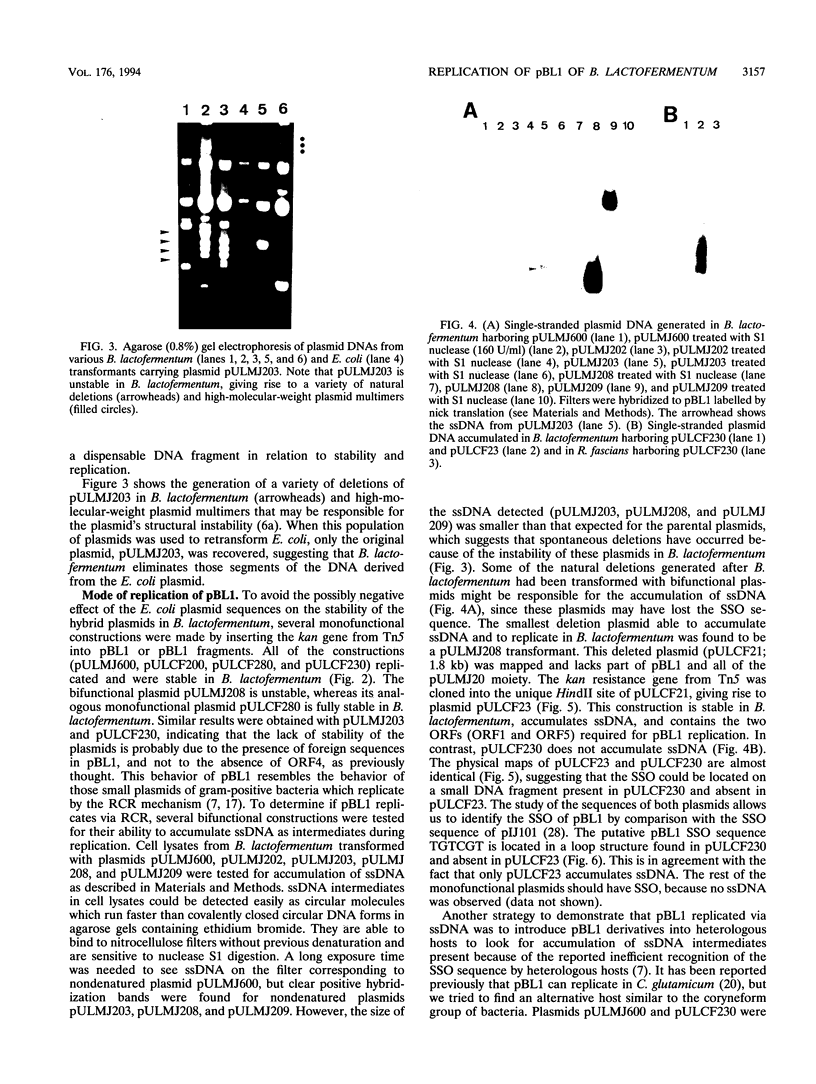
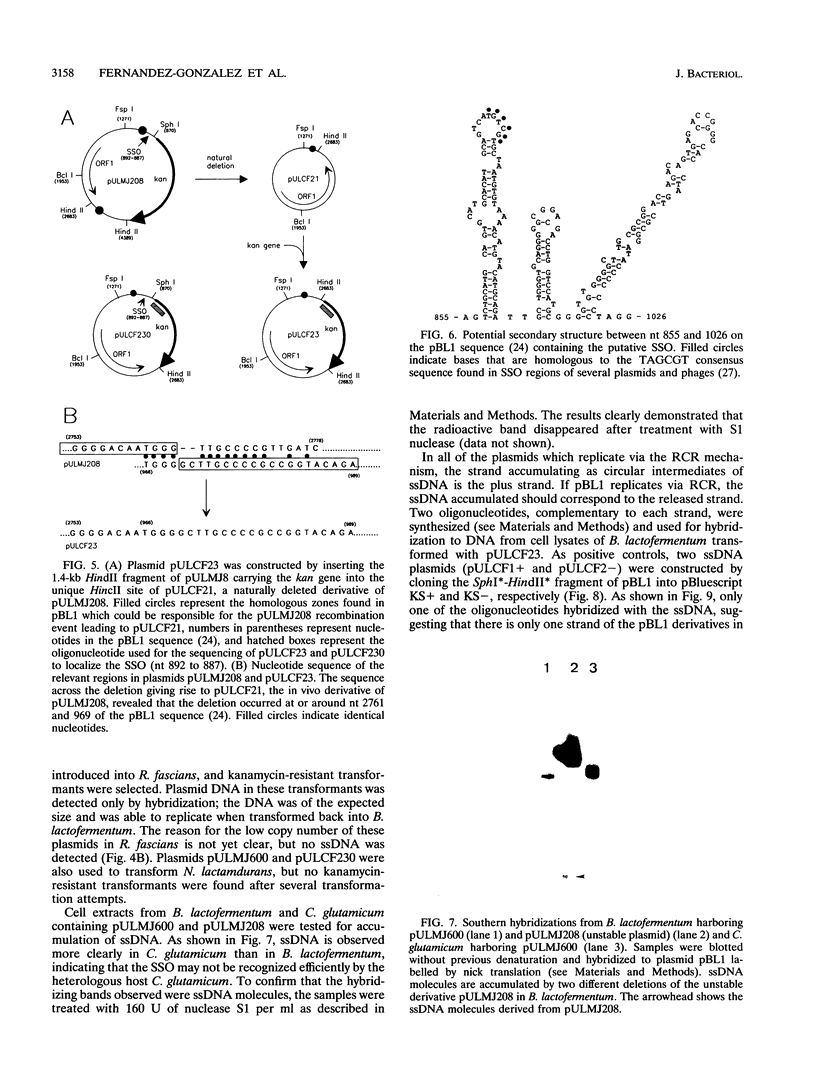
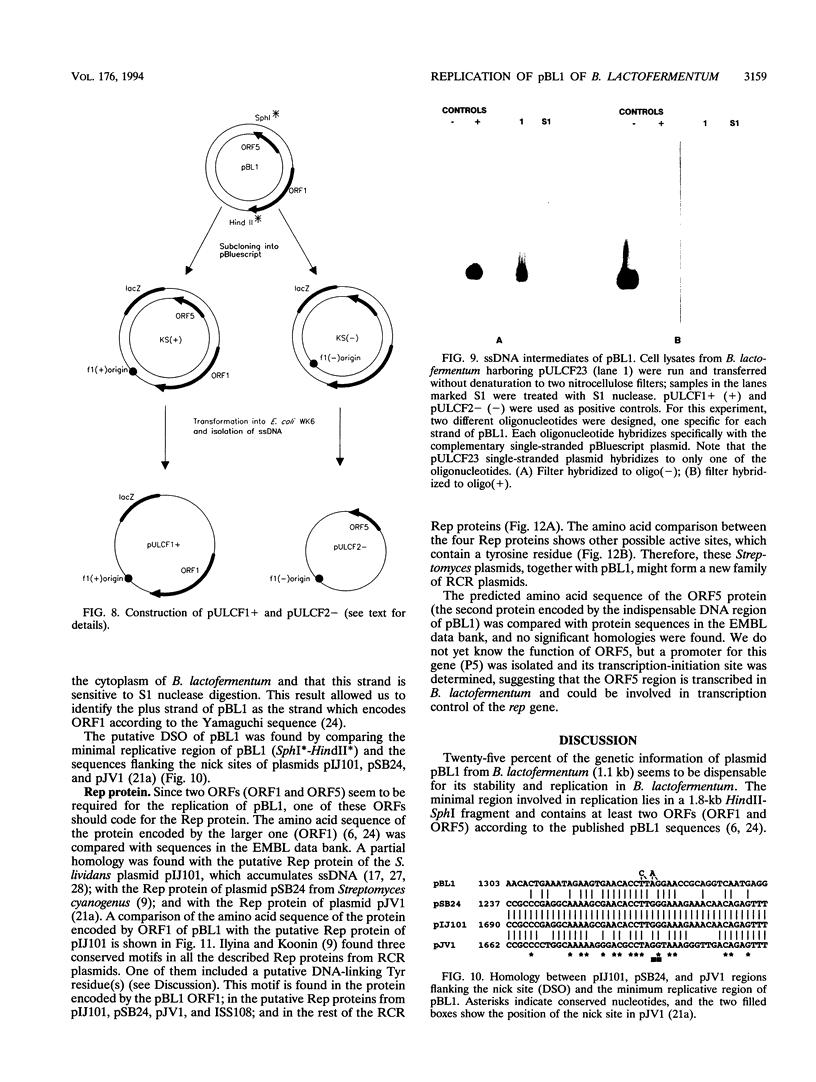
The minimal region for autonomous replication of pBL1, a 4.5-kb cryptic plasmid of Brevibacterium lactofermentum ATCC 13869 that has been used to construct a variety of corynebacterium vectors, was shown to be contained on a 1.8-kb HindII-SphI DNA fragment. This region contains two open reading frames (ORFs) (ORF1 and ORF5) which are essential for pBL1 replication in B. lactofermentum. Accumulation of single-strand intermediates in some of the constructions indicates that plasmid pBL1 replicates via the rolling circle replication model; its plus strand and minus strand were identified by hybridization with two synthetic oligonucleotide probes complementary to each pBL1 strand. ORF1 seems to encode the Rep protein and showed partial homology with sequences for Rep proteins from Streptomyces plasmids which replicate via rolling circle replication such as pIJ101, pSB24, and pJV1.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Albertini A. M., Hofer M., Calos M. P., Miller J. H. On the formation of spontaneous deletions: the importance of short sequence homologies in the generation of large deletions. Cell. 1982 Jun;29(2):319–328. doi: 10.1016/0092-8674(82)90148-9. [DOI] [PubMed] [Google Scholar]
- Archer J. A., Sinskey A. J. The DNA sequence and minimal replicon of the Corynebacterium glutamicum plasmid pSR1: evidence of a common ancestry with plasmids from C. diphtheriae. J Gen Microbiol. 1993 Aug;139(8):1753–1759. doi: 10.1099/00221287-139-8-1753. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cadenas R. F., Gil J. A., Martín J. F. Expression of Streptomyces genes encoding extracellular enzymes in Brevibacterium lactofermentum: secretion proceeds by removal of the same leader peptide as in Streptomyces lividans. Appl Microbiol Biotechnol. 1992 Dec;38(3):362–369. doi: 10.1007/BF00170087. [DOI] [PubMed] [Google Scholar]
- Cadenas R. F., Martín J. F., Gil J. A. Construction and characterization of promoter-probe vectors for Corynebacteria using the kanamycin-resistance reporter gene. Gene. 1991 Feb 1;98(1):117–121. doi: 10.1016/0378-1119(91)90113-p. [DOI] [PubMed] [Google Scholar]
- Crespi M., Messens E., Caplan A. B., van Montagu M., Desomer J. Fasciation induction by the phytopathogen Rhodococcus fascians depends upon a linear plasmid encoding a cytokinin synthase gene. EMBO J. 1992 Mar;11(3):795–804. doi: 10.1002/j.1460-2075.1992.tb05116.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. Insertion of foreign DNA into plasmids from gram-positive bacteria induces formation of high-molecular-weight plasmid multimers. J Bacteriol. 1988 Mar;170(3):1183–1190. doi: 10.1128/jb.170.3.1183-1190.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gruss A., Ehrlich S. D. The family of highly interrelated single-stranded deoxyribonucleic acid plasmids. Microbiol Rev. 1989 Jun;53(2):231–241. doi: 10.1128/mr.53.2.231-241.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina T. V., Koonin E. V. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992 Jul 11;20(13):3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen G. R., Bibb M. J. Derivatives of pUC18 that have BglII sites flanking a modified multiple cloning site and that retain the ability to identify recombinant clones by visual screening of Escherichia coli colonies. Gene. 1993 Feb 14;124(1):133–134. doi: 10.1016/0378-1119(93)90774-w. [DOI] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Novick R. P. Staphylococcal plasmids and their replication. Annu Rev Microbiol. 1989;43:537–565. doi: 10.1146/annurev.mi.43.100189.002541. [DOI] [PubMed] [Google Scholar]
- Pigac J., Vujaklija D., Toman Z., Gamulin V., Schrempf H. Structural instability of a bifunctional plasmid pZG1 and single-stranded DNA formation in Streptomyces. Plasmid. 1988 May;19(3):222–230. doi: 10.1016/0147-619x(88)90040-6. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaria R. I., Gil J. A., Martin J. F. High-frequency transformation of Brevibacterium lactofermentum protoplasts by plasmid DNA. J Bacteriol. 1985 Apr;162(1):463–467. doi: 10.1128/jb.162.1.463-467.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santamaría R. I., Martín J. F., Gil J. A. Identification of a promoter sequence in the plasmid pUL340 of Brevibacterium lactofermentum and construction of new cloning vectors for corynebacteria containing two selectable markers. Gene. 1987;56(2-3):199–208. doi: 10.1016/0378-1119(87)90137-5. [DOI] [PubMed] [Google Scholar]
- Servín-González L. Relationship between the replication functions of Streptomyces plasmids pJV1 and pIJ101. Plasmid. 1993 Sep;30(2):131–140. doi: 10.1006/plas.1993.1040. [DOI] [PubMed] [Google Scholar]
- Smith M. D., Flickinger J. L., Lineberger D. W., Schmidt B. Protoplast transformation in coryneform bacteria and introduction of an alpha-amylase gene from Bacillus amyloliquefaciens into Brevibacterium lactofermentum. Appl Environ Microbiol. 1986 Mar;51(3):634–639. doi: 10.1128/aem.51.3.634-639.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trautwetter A., Blanco C. Structural organization of the Corynebacterium glutamicum plasmid pCG100. J Gen Microbiol. 1991 Sep;137(9):2093–2101. doi: 10.1099/00221287-137-9-2093. [DOI] [PubMed] [Google Scholar]
- Yeh P., Oreglia J., Prévots F., Sicard A. M. A shuttle vector system for Brevibacterium lactofermentum. Gene. 1986;47(2-3):301–306. doi: 10.1016/0378-1119(86)90074-0. [DOI] [PubMed] [Google Scholar]
- Yoshihama M., Higashiro K., Rao E. A., Akedo M., Shanabruch W. G., Follettie M. T., Walker G. C., Sinskey A. J. Cloning vector system for Corynebacterium glutamicum. J Bacteriol. 1985 May;162(2):591–597. doi: 10.1128/jb.162.2.591-597.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaman S., Radnedge L., Richards H., Ward J. M. Analysis of the site for second-strand initiation during replication of the Streptomyces plasmid pIJ101. J Gen Microbiol. 1993 Apr;139(4):669–676. doi: 10.1099/00221287-139-4-669. [DOI] [PubMed] [Google Scholar]
- Zaman S., Richards H., Ward J. Identification of the minimal replicon of the streptomycete plasmid pIJ101. Plasmid. 1993 Jan;29(1):57–62. doi: 10.1006/plas.1993.1007. [DOI] [PubMed] [Google Scholar]