Abstract
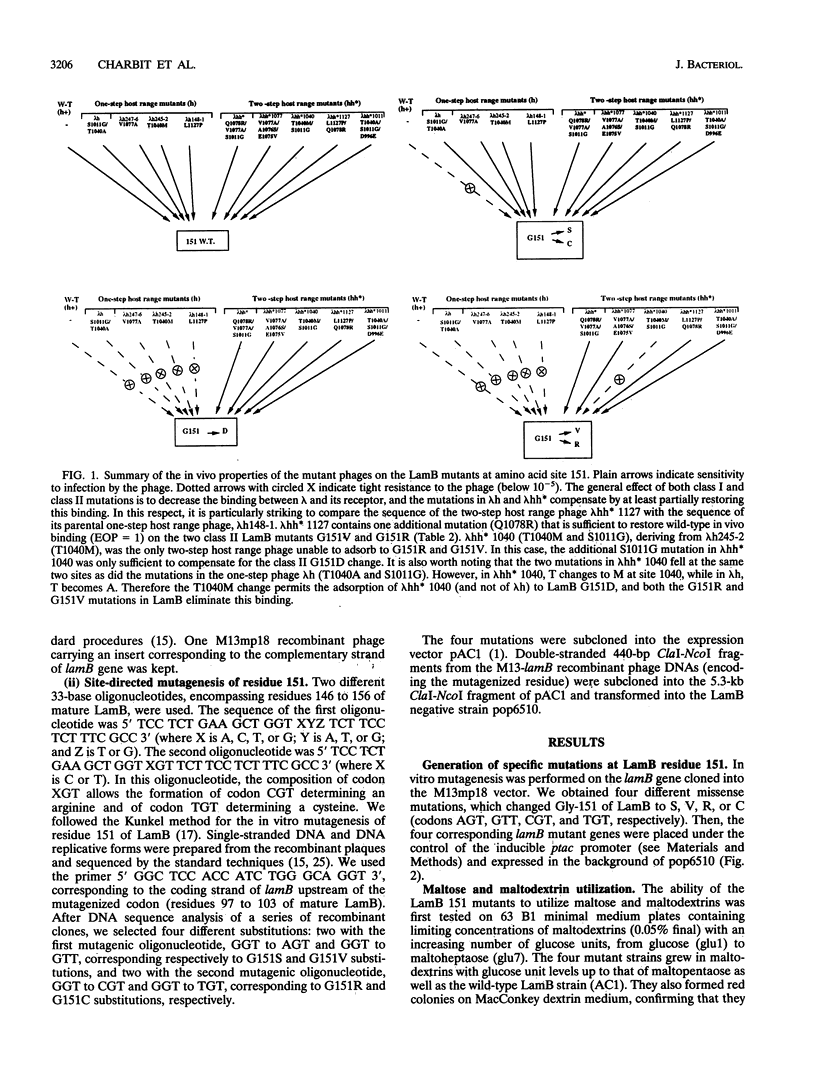
LamB is the cell surface receptor for bacteriophage lambda. LamB missense mutations yielding resistance to lambda have been previously grouped in two classes. Class I mutants block growth of lambda with wild-type host range (lambda h+) but support growth of one-step extended-host-range mutants (lambda h). Class II mutants block lambda h but support growth of two-step extended host range mutants (lambda hh*). While Class I mutations occur at 11 different amino acid sites, in five distinct portions of LamB, all the Class II mutations analyzed previously correspond to the same G-to-D change at amino acid 151. We generated by in vitro mutagenesis four different new substitutions at site 151 (to S, V, R, and C). Two of the mutants (G-151-->V [G151V] and G151R) were of Class II, while the two others (G151S and G151C) were of Class I, demonstrating that not only the site but also the nature of the substitutions at residue 151 was critical for the phage sensitivity phenotypes. The introduction of a negatively charged, a positively charged, or an aliphatic nonpolar residue at site 151 of LamB prevented both lambda h+ and lambda h adsorption, indicating that the block is not due to a charge effect. In contrast to G151D, which was sensitive to all the lambda hh* phages, G151V and G151R conferred sensitivity to only four of the five lambda hh* phages. Thus, G151V and G151R represent a new subclass of Class II LamB mutations that is more restrictive with respect to the growth of lambda hh*. Our results agree with the hypothesis that residue 151 belongs to an accessibility gate controlling the access to the phage tight-binding site and that substitutions at this residue affect the access of the phage to the binding site in relation to the size of the substitute side chain (surface area): the most restrictive changes are G151V and G151R, followed to a lesser extent by G151D and they by G151S and G151C.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boulain J. C., Charbit A., Hofnung M. Mutagenesis by random linker insertion into the lamB gene of Escherichia coli K12. Mol Gen Genet. 1986 Nov;205(2):339–348. doi: 10.1007/BF00430448. [DOI] [PubMed] [Google Scholar]
- Braun-Breton C., Hofnung M. In vivo and in vitro functional alterations of the bacteriophage lambda receptor in lamB missense mutants of Escherichia coli K-12. J Bacteriol. 1981 Dec;148(3):845–852. doi: 10.1128/jb.148.3.845-852.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chapon C. Role of the catabolite activator protein in the maltose regulon of Escherichia coli. J Bacteriol. 1982 May;150(2):722–729. doi: 10.1128/jb.150.2.722-729.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbit A., Clement J. M., Hofnung M. Further sequence analysis of the phage lambda receptor site. Possible implications for the organization of the lamB protein in Escherichia coli K12. J Mol Biol. 1984 May 25;175(3):395–401. doi: 10.1016/0022-2836(84)90355-3. [DOI] [PubMed] [Google Scholar]
- Charbit A., Gehring K., Nikaido H., Ferenci T., Hofnung M. Maltose transport and starch binding in phage-resistant point mutants of maltoporin. Functional and topological implications. J Mol Biol. 1988 Jun 5;201(3):487–496. doi: 10.1016/0022-2836(88)90630-4. [DOI] [PubMed] [Google Scholar]
- Charbit A., Hofnung M. Isolation of different bacteriophages using the LamB protein for adsorption on Escherichia coli K-12. J Virol. 1985 Feb;53(2):667–671. doi: 10.1128/jvi.53.2.667-671.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Charbit A., Ronco J., Michel V., Werts C., Hofnung M. Permissive sites and topology of an outer membrane protein with a reporter epitope. J Bacteriol. 1991 Jan;173(1):262–275. doi: 10.1128/jb.173.1.262-275.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chothia C. The nature of the accessible and buried surfaces in proteins. J Mol Biol. 1976 Jul 25;105(1):1–12. doi: 10.1016/0022-2836(76)90191-1. [DOI] [PubMed] [Google Scholar]
- Clément J. M., Hofnung M. Gene sequence of the lambda receptor, an outer membrane protein of E. coli K12. Cell. 1981 Dec;27(3 Pt 2):507–514. doi: 10.1016/0092-8674(81)90392-5. [DOI] [PubMed] [Google Scholar]
- Clément J. M., Lepouce E., Marchal C., Hofnung M. Genetic study of a membrane protein: DNA sequence alterations due to 17 lamB point mutations affecting adsorption of phage lambda. EMBO J. 1983;2(1):77–80. doi: 10.1002/j.1460-2075.1983.tb01384.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferenci T., Boos W. The role of the Escherichia coli lambda receptor in the transport of maltose and maltodextrins. J Supramol Struct. 1980;13(1):101–116. doi: 10.1002/jss.400130110. [DOI] [PubMed] [Google Scholar]
- Francis G., Brennan L., Stretton S., Ferenci T. Genetic mapping of starch- and lambda-receptor sites in maltoporin: identification of substitutions causing direct and indirect effects on binding sites by cysteine mutagenesis. Mol Microbiol. 1991 Sep;5(9):2293–2301. doi: 10.1111/j.1365-2958.1991.tb02160.x. [DOI] [PubMed] [Google Scholar]
- Gehring K., Charbit A., Brissaud E., Hofnung M. Bacteriophage lambda receptor site on the Escherichia coli K-12 LamB protein. J Bacteriol. 1987 May;169(5):2103–2106. doi: 10.1128/jb.169.5.2103-2106.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofnung M., Jezierska A., Braun-Breton C. lamB mutations in E. coli K12: growth of lambda host range mutants and effect of nonsense suppressors. Mol Gen Genet. 1976 May 7;145(2):207–213. doi: 10.1007/BF00269595. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marchal C., Hofnung M. Negative dominance in gene lamB: random assembly of secreted subunits issued from different polysomes. EMBO J. 1983;2(1):81–86. doi: 10.1002/j.1460-2075.1983.tb01385.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakae T., Ishii J. N. Molecular weights and subunit structure of LamB proteins. Ann Microbiol (Paris) 1982 Jan;133A(1):21–25. [PubMed] [Google Scholar]
- Neuhaus J. M. The receptor protein of phage lambda: purification, characterization and preliminary electrical studies in planar lipid bilayers. Ann Microbiol (Paris) 1982 Jan;133A(1):27–32. [PubMed] [Google Scholar]
- Randall-Hazelbauer L., Schwartz M. Isolation of the bacteriophage lambda receptor from Escherichia coli. J Bacteriol. 1973 Dec;116(3):1436–1446. doi: 10.1128/jb.116.3.1436-1446.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rees D. C., DeAntonio L., Eisenberg D. Hydrophobic organization of membrane proteins. Science. 1989 Aug 4;245(4917):510–513. doi: 10.1126/science.2667138. [DOI] [PubMed] [Google Scholar]
- Roa M., Clément J. M. Location of a phage binding region on an outer membrane protein. FEBS Lett. 1980 Nov 17;121(1):127–129. doi: 10.1016/0014-5793(80)81280-4. [DOI] [PubMed] [Google Scholar]
- Roa M. Interaction of bacteriophage K10 with its receptor, the lamB protein of Escherichia coli. J Bacteriol. 1979 Nov;140(2):680–686. doi: 10.1128/jb.140.2.680-686.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schirmer T., Cowan S. W. Prediction of membrane-spanning beta-strands and its application to maltoporin. Protein Sci. 1993 Aug;2(8):1361–1363. doi: 10.1002/pro.5560020820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szmelcman S., Hofnung M. Maltose transport in Escherichia coli K-12: involvement of the bacteriophage lambda receptor. J Bacteriol. 1975 Oct;124(1):112–118. doi: 10.1128/jb.124.1.112-118.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thirion J. P., Hofnung M. On some genetic aspects of phage lambda resistance in E. coli K12. Genetics. 1972 Jun;71(2):207–216. doi: 10.1093/genetics/71.2.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werts C., Charbit A. Construction and first characterization of two reciprocal hybrids between LamB from Escherichia coli K12 and Klebsiella pneumoniae. Res Microbiol. 1993 May;144(4):259–269. doi: 10.1016/0923-2508(93)90010-y. [DOI] [PubMed] [Google Scholar]
- Werts C., Michel V., Hofnung M., Charbit A. Adsorption of bacteriophage lambda on the LamB protein of Escherichia coli K-12: point mutations in gene J of lambda responsible for extended host range. J Bacteriol. 1994 Feb;176(4):941–947. doi: 10.1128/jb.176.4.941-947.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werts C., O'Callaghan D., Hofnung M., Charbit A. Immunological relatedness of the LamB proteins among members of Enterobacteriaceae. J Gen Microbiol. 1993 Apr;139(4):881–887. doi: 10.1099/00221287-139-4-881. [DOI] [PubMed] [Google Scholar]