Abstract
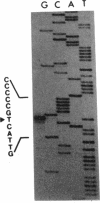
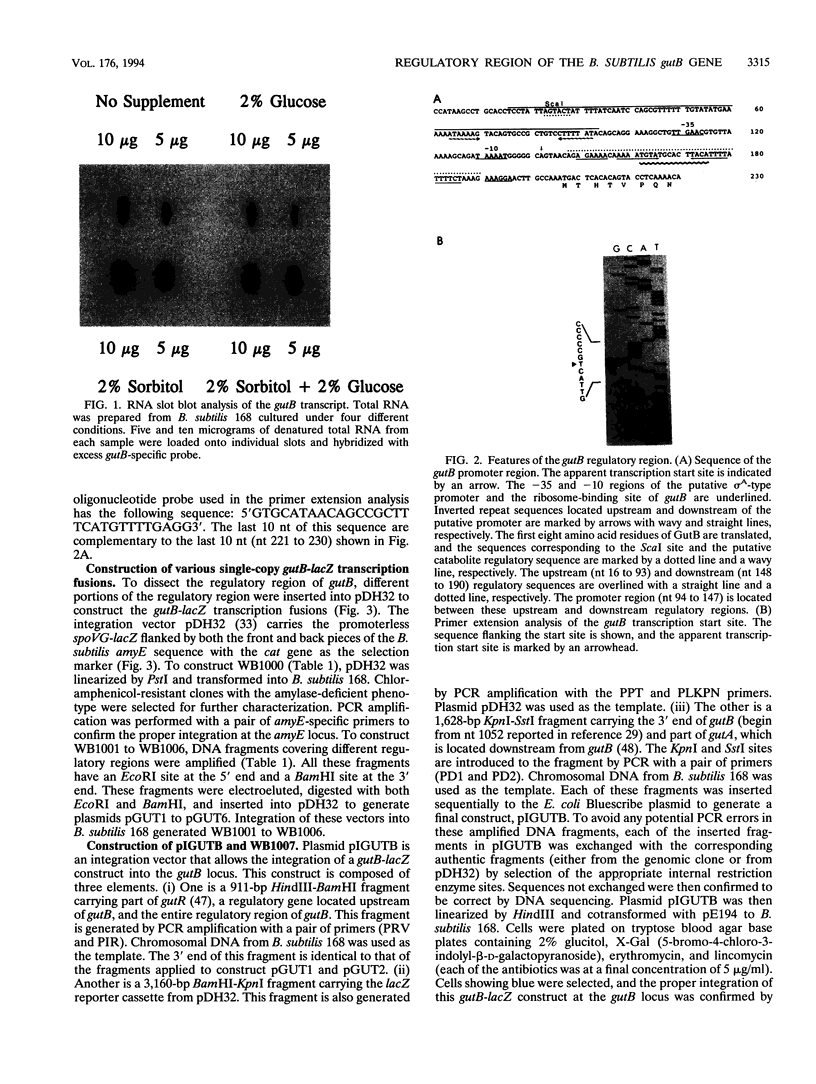
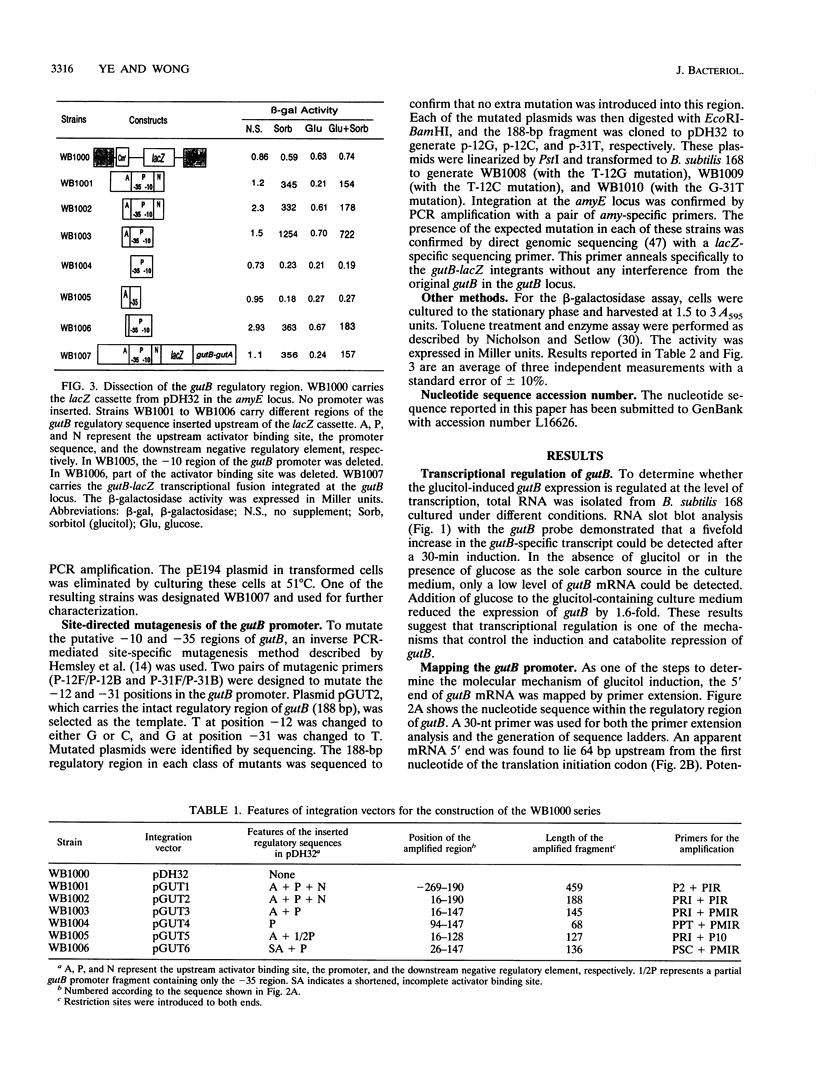
The regulatory region of the Bacillus subtilis glucitol dehydrogenase (gutB) gene was divided into three subregions: a promoter, an upstream positive regulatory region, and a downstream negative regulatory region. Data from primer extension, deletion, and site-directed mutagenesis analyses were consistent with two possible models for the gutB promoter. It is either a sigma A-type promoter with an unusually short spacer region (15 bp) or a special sigma A promoter which requires only the hexameric -10 sequence for its function. Sequence carrying just the promoter region (from -48 to +6) failed to direct transcription in vivo. An upstream regulatory sequence was essential for glucitol induction. When this sequence was inserted in a high-copy-number plasmid, an effect characteristic of titration of a transcriptional activator was seen. Downstream from the promoter, there is an imperfect, AT-rich inverted repeat sequence. Deletion of this element did not lead to constitutive expression of gutB. However, the induced gutB expression level was enhanced three- to fourfold.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anagnostopoulos C., Spizizen J. REQUIREMENTS FOR TRANSFORMATION IN BACILLUS SUBTILIS. J Bacteriol. 1961 May;81(5):741–746. doi: 10.1128/jb.81.5.741-746.1961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalumeau H., Delobbe A., Gay P. Biochemical and genetic study of D-glucitol transport and catabolism in Bacillus subtilis. J Bacteriol. 1978 Jun;134(3):920–928. doi: 10.1128/jb.134.3.920-928.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delobbe A., Chalumeau H., Gay P. Existence of two alternative pathways for fructose and sorbitol metabolism in Bacillus subtilis Marburg. Eur J Biochem. 1975 Feb 21;51(2):503–510. doi: 10.1111/j.1432-1033.1975.tb03950.x. [DOI] [PubMed] [Google Scholar]
- Débarbouillé M., Martin-Verstraete I., Kunst F., Rapoport G. The Bacillus subtilis sigL gene encodes an equivalent of sigma 54 from gram-negative bacteria. Proc Natl Acad Sci U S A. 1991 Oct 15;88(20):9092–9096. doi: 10.1073/pnas.88.20.9092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emory S. A., Bouvet P., Belasco J. G. A 5'-terminal stem-loop structure can stabilize mRNA in Escherichia coli. Genes Dev. 1992 Jan;6(1):135–148. doi: 10.1101/gad.6.1.135. [DOI] [PubMed] [Google Scholar]
- Fisher S. H., Sonenshein A. L. Control of carbon and nitrogen metabolism in Bacillus subtilis. Annu Rev Microbiol. 1991;45:107–135. doi: 10.1146/annurev.mi.45.100191.000543. [DOI] [PubMed] [Google Scholar]
- Fouet A., Jin S. F., Raffel G., Sonenshein A. L. Multiple regulatory sites in the Bacillus subtilis citB promoter region. J Bacteriol. 1990 Sep;172(9):5408–5415. doi: 10.1128/jb.172.9.5408-5415.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gay P., Chalumeau H., Steinmetz M. Chromosomal localization of gut, fruC, and pfk mutations affecting genes involved in Bacillus subtilis D-glucitol catabolism. J Bacteriol. 1983 Mar;153(3):1133–1137. doi: 10.1128/jb.153.3.1133-1137.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilman M. Z., Wiggs J. L., Chamberlin M. J. Nucleotide sequences of two Bacillus subtilis promoters used by Bacillus subtilis sigma-28 RNA polymerase. Nucleic Acids Res. 1981 Nov 25;9(22):5991–6000. doi: 10.1093/nar/9.22.5991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gärtner D., Geissendörfer M., Hillen W. Expression of the Bacillus subtilis xyl operon is repressed at the level of transcription and is induced by xylose. J Bacteriol. 1988 Jul;170(7):3102–3109. doi: 10.1128/jb.170.7.3102-3109.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helmann J. D., Wang Y., Mahler I., Walsh C. T. Homologous metalloregulatory proteins from both gram-positive and gram-negative bacteria control transcription of mercury resistance operons. J Bacteriol. 1989 Jan;171(1):222–229. doi: 10.1128/jb.171.1.222-229.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemsley A., Arnheim N., Toney M. D., Cortopassi G., Galas D. J. A simple method for site-directed mutagenesis using the polymerase chain reaction. Nucleic Acids Res. 1989 Aug 25;17(16):6545–6551. doi: 10.1093/nar/17.16.6545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoch J. A. The phosphorelay signal transduction pathway in the initiation of Bacillus subtilis sporulation. J Cell Biochem. 1993 Jan;51(1):55–61. doi: 10.1002/jcb.240510111. [DOI] [PubMed] [Google Scholar]
- Kenney T. J., Kirchman P. A., Moran C. P., Jr Gene encoding sigma E is transcribed from a sigma A-like promoter in Bacillus subtilis. J Bacteriol. 1988 Jul;170(7):3058–3064. doi: 10.1128/jb.170.7.3058-3064.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney T. J., Moran C. P., Jr Genetic evidence for interaction of sigma A with two promoters in Bacillus subtilis. J Bacteriol. 1991 Jun;173(11):3282–3290. doi: 10.1128/jb.173.11.3282-3290.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney T. J., York K., Youngman P., Moran C. P., Jr Genetic evidence that RNA polymerase associated with sigma A factor uses a sporulation-specific promoter in Bacillus subtilis. Proc Natl Acad Sci U S A. 1989 Dec;86(23):9109–9113. doi: 10.1073/pnas.86.23.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klier A. F., Rapoport G. Genetics and regulation of carbohydrate catabolism in Bacillus. Annu Rev Microbiol. 1988;42:65–95. doi: 10.1146/annurev.mi.42.100188.000433. [DOI] [PubMed] [Google Scholar]
- Klier A., Msadek T., Rapoport G. Positive regulation in the gram-positive bacterium: Bacillus subtilis. Annu Rev Microbiol. 1992;46:429–459. doi: 10.1146/annurev.mi.46.100192.002241. [DOI] [PubMed] [Google Scholar]
- Li M., Wong S. L. Cloning and characterization of the groESL operon from Bacillus subtilis. J Bacteriol. 1992 Jun;174(12):3981–3992. doi: 10.1128/jb.174.12.3981-3992.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez J. M., Uratani-Wong B., Freese E. Catabolite repression of enzyme synthesis does not prevent sporulation. J Bacteriol. 1980 Mar;141(3):1447–1449. doi: 10.1128/jb.141.3.1447-1449.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund P. A., Ford S. J., Brown N. L. Transcriptional regulation of the mercury-resistance genes of transposon Tn501. J Gen Microbiol. 1986 Feb;132(2):465–480. doi: 10.1099/00221287-132-2-465. [DOI] [PubMed] [Google Scholar]
- Martin-Verstraete I., Débarbouillé M., Klier A., Rapoport G. Levanase operon of Bacillus subtilis includes a fructose-specific phosphotransferase system regulating the expression of the operon. J Mol Biol. 1990 Aug 5;214(3):657–671. doi: 10.1016/0022-2836(90)90284-S. [DOI] [PubMed] [Google Scholar]
- Martin-Verstraete I., Débarbouillé M., Klier A., Rapoport G. Mutagenesis of the Bacillus subtilis "-12, -24" promoter of the levanase operon and evidence for the existence of an upstream activating sequence. J Mol Biol. 1992 Jul 5;226(1):85–99. doi: 10.1016/0022-2836(92)90126-5. [DOI] [PubMed] [Google Scholar]
- Melin L., Fridén H., Dehlin E., Rutberg L., von Gabain A. The importance of the 5'-region in regulating the stability of sdh mRNA in Bacillus subtilis. Mol Microbiol. 1990 Nov;4(11):1881–1889. doi: 10.1111/j.1365-2958.1990.tb02037.x. [DOI] [PubMed] [Google Scholar]
- Miwa Y., Fujita Y. Determination of the cis sequence involved in catabolite repression of the Bacillus subtilis gnt operon; implication of a consensus sequence in catabolite repression in the genus Bacillus. Nucleic Acids Res. 1990 Dec 11;18(23):7049–7053. doi: 10.1093/nar/18.23.7049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran C. P., Jr, Lang N., LeGrice S. F., Lee G., Stephens M., Sonenshein A. L., Pero J., Losick R. Nucleotide sequences that signal the initiation of transcription and translation in Bacillus subtilis. Mol Gen Genet. 1982;186(3):339–346. doi: 10.1007/BF00729452. [DOI] [PubMed] [Google Scholar]
- Ng K., Ye R., Wu X. C., Wong S. L. Sorbitol dehydrogenase from Bacillus subtilis. Purification, characterization, and gene cloning. J Biol Chem. 1992 Dec 15;267(35):24989–24994. [PubMed] [Google Scholar]
- O'Halloran T., Walsh C. Metalloregulatory DNA-binding protein encoded by the merR gene: isolation and characterization. Science. 1987 Jan 9;235(4785):211–214. doi: 10.1126/science.3798107. [DOI] [PubMed] [Google Scholar]
- Pang A. S., Nathoo S., Wong S. L. Cloning and characterization of a pair of novel genes that regulate production of extracellular enzymes in Bacillus subtilis. J Bacteriol. 1991 Jan;173(1):46–54. doi: 10.1128/jb.173.1.46-54.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reizer J., Romano A. H., Deutscher J. The role of phosphorylation of HPr, a phosphocarrier protein of the phosphotransferase system, in the regulation of carbon metabolism in gram-positive bacteria. J Cell Biochem. 1993 Jan;51(1):19–24. doi: 10.1002/jcb.240510105. [DOI] [PubMed] [Google Scholar]
- Satola S., Kirchman P. A., Moran C. P., Jr Spo0A binds to a promoter used by sigma A RNA polymerase during sporulation in Bacillus subtilis. Proc Natl Acad Sci U S A. 1991 May 15;88(10):4533–4537. doi: 10.1073/pnas.88.10.4533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shewchuk L. M., Verdine G. L., Walsh C. T. Transcriptional switching by the metalloregulatory MerR protein: initial characterization of DNA and mercury (II) binding activities. Biochemistry. 1989 Mar 7;28(5):2331–2339. doi: 10.1021/bi00431a052. [DOI] [PubMed] [Google Scholar]
- Shimotsu H., Henner D. J. Construction of a single-copy integration vector and its use in analysis of regulation of the trp operon of Bacillus subtilis. Gene. 1986;43(1-2):85–94. doi: 10.1016/0378-1119(86)90011-9. [DOI] [PubMed] [Google Scholar]
- Stewart G. C. Catabolite repression in the gram-positive bacteria: generation of negative regulators of transcription. J Cell Biochem. 1993 Jan;51(1):25–28. doi: 10.1002/jcb.240510106. [DOI] [PubMed] [Google Scholar]
- Wang L. F., Doi R. H. Promoter switching during development and the termination site of the sigma 43 operon of Bacillus subtilis. Mol Gen Genet. 1987 Apr;207(1):114–119. doi: 10.1007/BF00331498. [DOI] [PubMed] [Google Scholar]
- Weickert M. J., Chambliss G. H. Site-directed mutagenesis of a catabolite repression operator sequence in Bacillus subtilis. Proc Natl Acad Sci U S A. 1990 Aug;87(16):6238–6242. doi: 10.1073/pnas.87.16.6238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong S. L. Development of an inducible and enhancible expression and secretion system in Bacillus subtilis. Gene. 1989 Nov 30;83(2):215–223. doi: 10.1016/0378-1119(89)90107-8. [DOI] [PubMed] [Google Scholar]
- Wong S. L., Wang L. F., Doi R. H. Cloning and nucleotide sequence of senN, a novel 'Bacillus natto' (B. subtilis) gene that regulates expression of extracellular protein genes. J Gen Microbiol. 1988 Dec;134(12):3269–3276. doi: 10.1099/00221287-134-12-3269. [DOI] [PubMed] [Google Scholar]
- Ye R., Rehemtulla S. N., Wong S. L. Glucitol induction in Bacillus subtilis is mediated by a regulatory factor, GutR. J Bacteriol. 1994 Jun;176(11):3321–3327. doi: 10.1128/jb.176.11.3321-3327.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- York K., Kenney T. J., Satola S., Moran C. P., Jr, Poth H., Youngman P. Spo0A controls the sigma A-dependent activation of Bacillus subtilis sporulation-specific transcription unit spoIIE. J Bacteriol. 1992 Apr;174(8):2648–2658. doi: 10.1128/jb.174.8.2648-2658.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Ham S. M., van Alphen L., Mooi F. R., van Putten J. P. Phase variation of H. influenzae fimbriae: transcriptional control of two divergent genes through a variable combined promoter region. Cell. 1993 Jun 18;73(6):1187–1196. doi: 10.1016/0092-8674(93)90647-9. [DOI] [PubMed] [Google Scholar]