Abstract
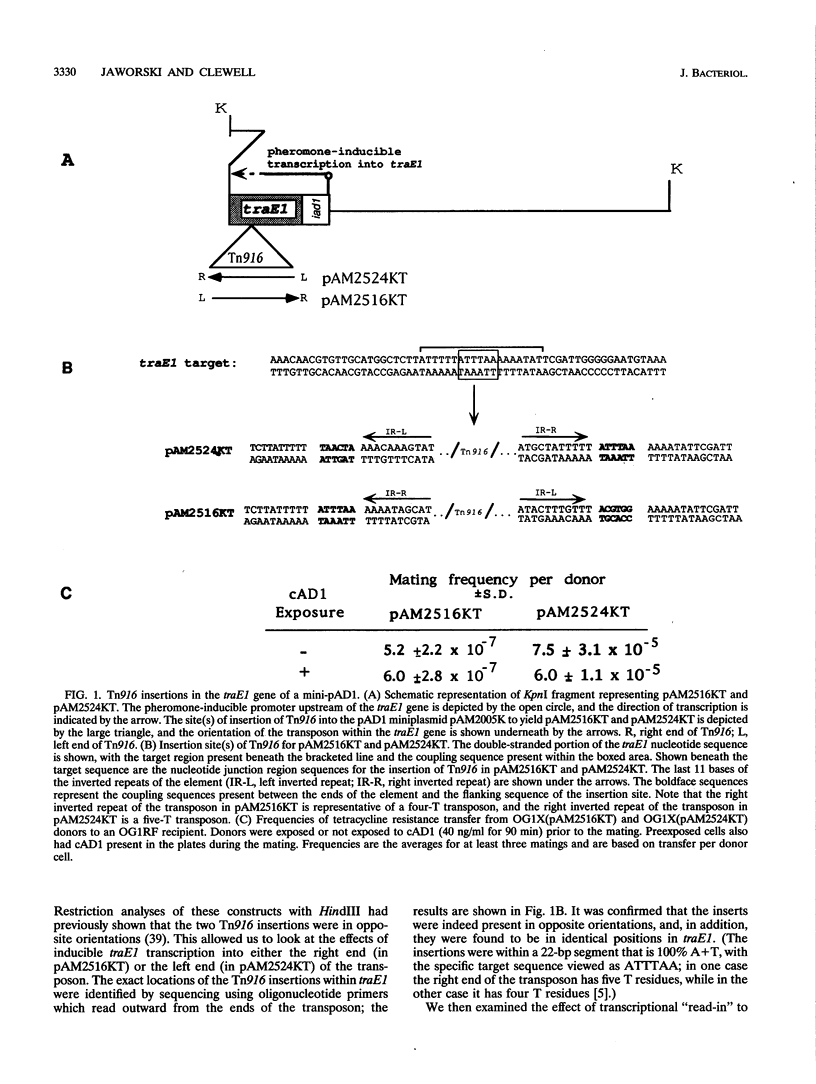
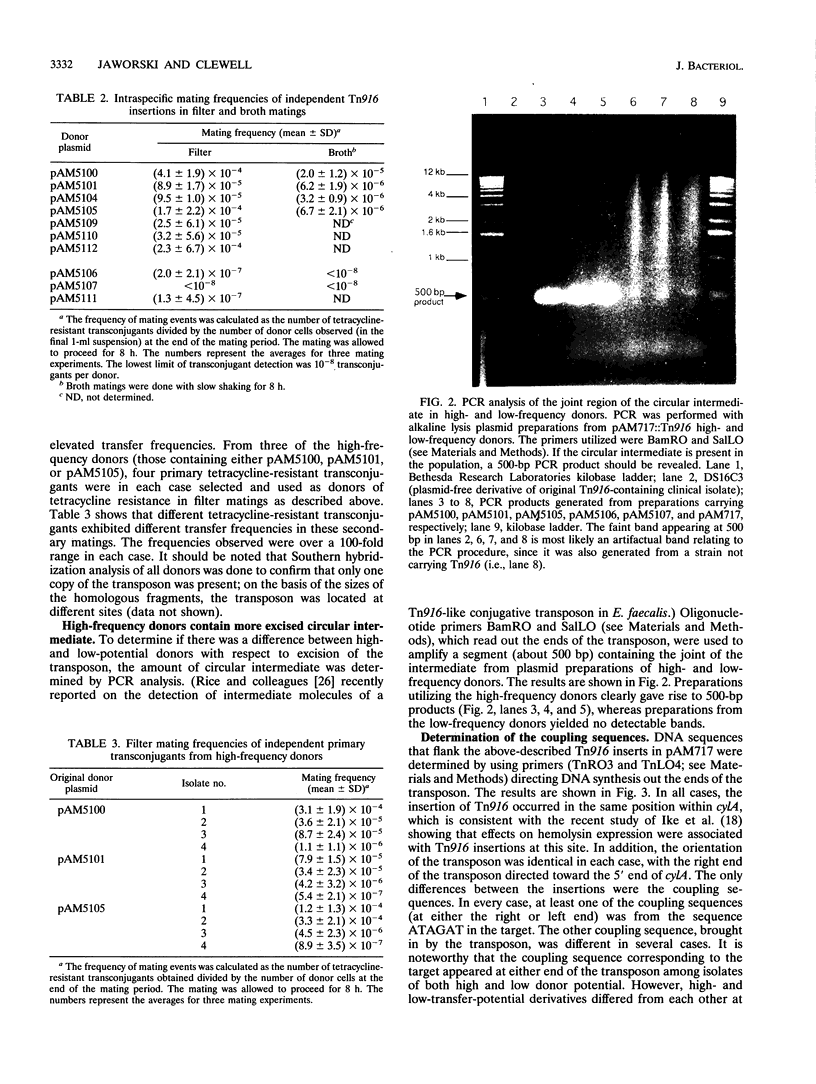
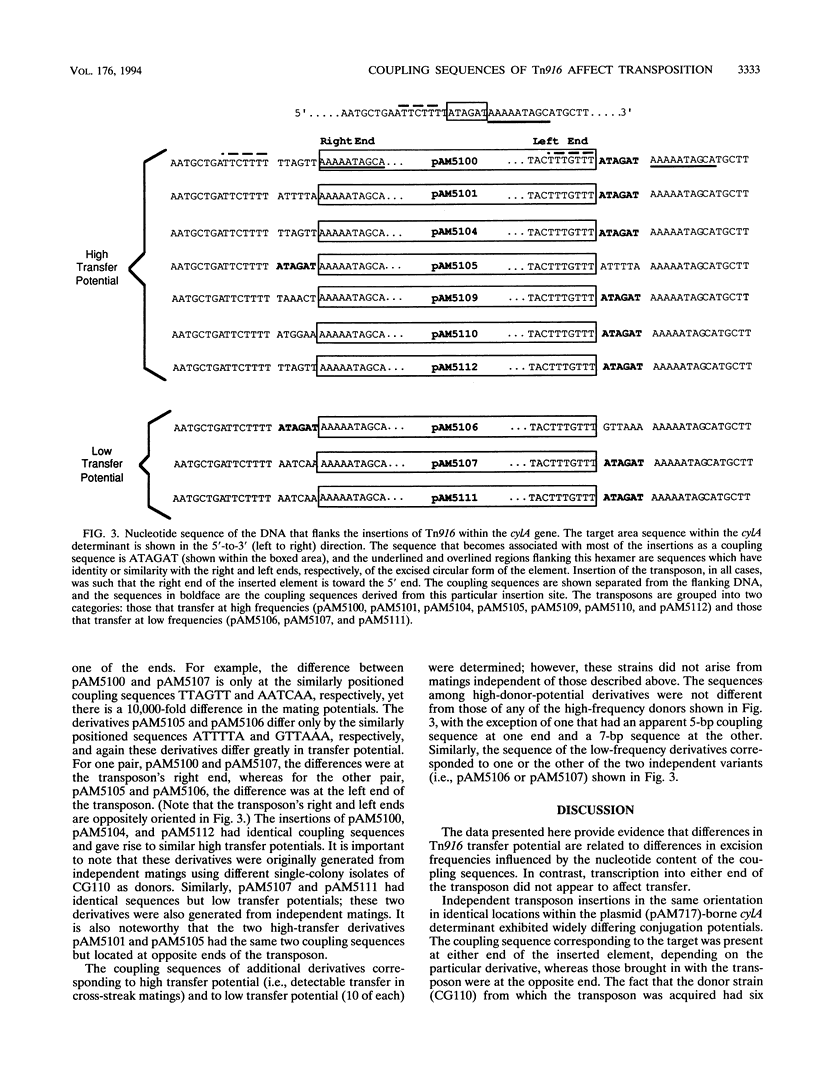
The conjugative transposon Tn916 (encodes resistance to tetracycline), originally identified in Enterococcus faecalis, moves by an excision-insertion process in which the rate-limiting step is believed to be excision. Individual transposon-containing strains exhibit characteristic mating frequencies which range over several orders of magnitude; the basis of this phenomenon is addressed in the present study. We were able to generate independent single-copy insertions in identical target locations and with similar orientations within a plasmid hemolysin determinant (cylA); however, transposition from this site occurred at very different frequencies (10(-8) to 10(-4) per donor) depending on the individual isolate. DNA sequencing analyses showed that the coupling (junction) sequences differed between isolates and thus appeared to be responsible for differences in excision frequencies. Other experiments showed that inducible transcription into either end of the transposon had no significant effect on transfer.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bringel F., Van Alstine G. L., Scott J. R. Transfer of Tn916 between Lactococcus lactis subsp. lactis strains is nontranspositional: evidence for a chromosomal fertility function in strain MG1363. J Bacteriol. 1992 Sep;174(18):5840–5847. doi: 10.1128/jb.174.18.5840-5847.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud F., Courvalin P. Nucleotide sequence of the ends of the conjugative shuttle transposon Tn1545. Mol Gen Genet. 1987 Aug;209(1):110–115. doi: 10.1007/BF00329844. [DOI] [PubMed] [Google Scholar]
- Caparon M. G., Scott J. R. Excision and insertion of the conjugative transposon Tn916 involves a novel recombination mechanism. Cell. 1989 Dec 22;59(6):1027–1034. doi: 10.1016/0092-8674(89)90759-9. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., An F. Y., White B. A., Gawron-Burke C. Streptococcus faecalis sex pheromone (cAM373) also produced by Staphylococcus aureus and identification of a conjugative transposon (Tn918). J Bacteriol. 1985 Jun;162(3):1212–1220. doi: 10.1128/jb.162.3.1212-1220.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Flannagan S. E., Ike Y., Jones J. M., Gawron-Burke C. Sequence analysis of termini of conjugative transposon Tn916. J Bacteriol. 1988 Jul;170(7):3046–3052. doi: 10.1128/jb.170.7.3046-3052.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clewell D. B., Gawron-Burke C. Conjugative transposons and the dissemination of antibiotic resistance in streptococci. Annu Rev Microbiol. 1986;40:635–659. doi: 10.1146/annurev.mi.40.100186.003223. [DOI] [PubMed] [Google Scholar]
- Clewell D. B., Tomich P. K., Gawron-Burke M. C., Franke A. E., Yagi Y., An F. Y. Mapping of Streptococcus faecalis plasmids pAD1 and pAD2 and studies relating to transposition of Tn917. J Bacteriol. 1982 Dec;152(3):1220–1230. doi: 10.1128/jb.152.3.1220-1230.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courvalin P., Carlier C. Transposable multiple antibiotic resistance in Streptococcus pneumoniae. Mol Gen Genet. 1986 Nov;205(2):291–297. doi: 10.1007/BF00430441. [DOI] [PubMed] [Google Scholar]
- Cruz-Rodz A. L., Gilmore M. S. High efficiency introduction of plasmid DNA into glycine treated Enterococcus faecalis by electroporation. Mol Gen Genet. 1990 Oct;224(1):152–154. doi: 10.1007/BF00259462. [DOI] [PubMed] [Google Scholar]
- Flannagan S. E., Clewell D. B. Conjugative transfer of Tn916 in Enterococcus faecalis: trans activation of homologous transposons. J Bacteriol. 1991 Nov;173(22):7136–7141. doi: 10.1128/jb.173.22.7136-7141.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franke A. E., Clewell D. B. Evidence for a chromosome-borne resistance transposon (Tn916) in Streptococcus faecalis that is capable of "conjugal" transfer in the absence of a conjugative plasmid. J Bacteriol. 1981 Jan;145(1):494–502. doi: 10.1128/jb.145.1.494-502.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. A transposon in Streptococcus faecalis with fertility properties. Nature. 1982 Nov 18;300(5889):281–284. doi: 10.1038/300281a0. [DOI] [PubMed] [Google Scholar]
- Gawron-Burke C., Clewell D. B. Regeneration of insertionally inactivated streptococcal DNA fragments after excision of transposon Tn916 in Escherichia coli: strategy for targeting and cloning of genes from gram-positive bacteria. J Bacteriol. 1984 Jul;159(1):214–221. doi: 10.1128/jb.159.1.214-221.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike Y., Clewell D. B. Genetic analysis of the pAD1 pheromone response in Streptococcus faecalis, using transposon Tn917 as an insertional mutagen. J Bacteriol. 1984 Jun;158(3):777–783. doi: 10.1128/jb.158.3.777-783.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike Y., Craig R. A., White B. A., Yagi Y., Clewell D. B. Modification of Streptococcus faecalis sex pheromones after acquisition of plasmid DNA. Proc Natl Acad Sci U S A. 1983 Sep;80(17):5369–5373. doi: 10.1073/pnas.80.17.5369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ike Y., Flannagan S. E., Clewell D. B. Hyperhemolytic phenomena associated with insertions of Tn916 into the hemolysin determinant of Enterococcus faecalis plasmid pAD1. J Bacteriol. 1992 Mar;174(6):1801–1809. doi: 10.1128/jb.174.6.1801-1809.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitts P. A., Nash H. A. Homology-dependent interactions in phage lambda site-specific recombination. Nature. 1987 Sep 24;329(6137):346–348. doi: 10.1038/329346a0. [DOI] [PubMed] [Google Scholar]
- Landy A. Dynamic, structural, and regulatory aspects of lambda site-specific recombination. Annu Rev Biochem. 1989;58:913–949. doi: 10.1146/annurev.bi.58.070189.004405. [DOI] [PubMed] [Google Scholar]
- Landy A., Hoess R. H., Bidwell K., Ross W. Site-specific recombination in bacteriophage lambda: structural features of recombining sites. Cold Spring Harb Symp Quant Biol. 1979;43(Pt 2):1089–1097. doi: 10.1101/sqb.1979.043.01.121. [DOI] [PubMed] [Google Scholar]
- Oliver D. R., Brown B. L., Clewell D. B. Analysis of plasmid deoxyribonucleic acid in a cariogenic strain of Streptococcus faecalis: an approach to identifying genetic determinants on cryptic plasmids. J Bacteriol. 1977 May;130(2):759–765. doi: 10.1128/jb.130.2.759-765.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pontius L. T., Clewell D. B. Conjugative transfer of Enterococcus faecalis plasmid pAD1: nucleotide sequence and transcriptional fusion analysis of a region involved in positive regulation. J Bacteriol. 1992 May;174(10):3152–3160. doi: 10.1128/jb.174.10.3152-3160.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart-Salmeron C., Trieu-Cuot P., Carlier C., Courvalin P. Molecular characterization of two proteins involved in the excision of the conjugative transposon Tn1545: homologies with other site-specific recombinases. EMBO J. 1989 Aug;8(8):2425–2433. doi: 10.1002/j.1460-2075.1989.tb08373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poyart-Salmeron C., Trieu-Cuot P., Carlier C., Courvalin P. The integration-excision system of the conjugative transposon Tn 1545 is structurally and functionally related to those of lambdoid phages. Mol Microbiol. 1990 Sep;4(9):1513–1521. doi: 10.1111/j.1365-2958.1990.tb02062.x. [DOI] [PubMed] [Google Scholar]
- Rice L. B., Marshall S. H., Carias L. L. Tn5381, a conjugative transposon identifiable as a circular form in Enterococcus faecalis. J Bacteriol. 1992 Nov;174(22):7308–7315. doi: 10.1128/jb.174.22.7308-7315.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. R., Kirchman P. A., Caparon M. G. An intermediate in transposition of the conjugative transposon Tn916. Proc Natl Acad Sci U S A. 1988 Jul;85(13):4809–4813. doi: 10.1073/pnas.85.13.4809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott J. R. Sex and the single circle: conjugative transposition. J Bacteriol. 1992 Oct;174(19):6005–6010. doi: 10.1128/jb.174.19.6005-6010.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Senghas E., Jones J. M., Yamamoto M., Gawron-Burke C., Clewell D. B. Genetic organization of the bacterial conjugative transposon Tn916. J Bacteriol. 1988 Jan;170(1):245–249. doi: 10.1128/jb.170.1.245-249.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimada K., Weisberg R. A., Gottesman M. E. Prophage lambda at unusual chromosomal locations. I. Location of the secondary attachment sites and the properties of the lysogens. J Mol Biol. 1972 Feb 14;63(3):483–503. doi: 10.1016/0022-2836(72)90443-3. [DOI] [PubMed] [Google Scholar]
- Su Y. A., Clewell D. B. Characterization of the left 4 kb of conjugative transposon Tn916: determinants involved in excision. Plasmid. 1993 Nov;30(3):234–250. doi: 10.1006/plas.1993.1055. [DOI] [PubMed] [Google Scholar]
- Su Y. A., Sulavik M. C., He P., Makinen K. K., Makinen P. L., Fiedler S., Wirth R., Clewell D. B. Nucleotide sequence of the gelatinase gene (gelE) from Enterococcus faecalis subsp. liquefaciens. Infect Immun. 1991 Jan;59(1):415–420. doi: 10.1128/iai.59.1.415-420.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanimoto K., Clewell D. B. Regulation of the pAD1-encoded sex pheromone response in Enterococcus faecalis: expression of the positive regulator TraE1. J Bacteriol. 1993 Feb;175(4):1008–1018. doi: 10.1128/jb.175.4.1008-1018.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson J. F., Landy A. Empirical estimation of protein-induced DNA bending angles: applications to lambda site-specific recombination complexes. Nucleic Acids Res. 1988 Oct 25;16(20):9687–9705. doi: 10.1093/nar/16.20.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trieu-Cuot P., Poyart-Salmeron C., Carlier C., Courvalin P. Sequence requirements for target activity in site-specific recombination mediated by the Int protein of transposon Tn 1545. Mol Microbiol. 1993 Apr;8(1):179–185. doi: 10.1111/j.1365-2958.1993.tb01214.x. [DOI] [PubMed] [Google Scholar]
- Weaver K. E., Clewell D. B. Construction of Enterococcus faecalis pAD1 miniplasmids: identification of a minimal pheromone response regulatory region and evaluation of a novel pheromone-dependent growth inhibition. Plasmid. 1989 Sep;22(2):106–119. doi: 10.1016/0147-619x(89)90020-6. [DOI] [PubMed] [Google Scholar]
- Weaver K. E., Clewell D. B. Regulation of the pAD1 sex pheromone response in Enterococcus faecalis: construction and characterization of lacZ transcriptional fusions in a key control region of the plasmid. J Bacteriol. 1988 Sep;170(9):4343–4352. doi: 10.1128/jb.170.9.4343-4352.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]