Abstract
The viral reverse transcriptase (RT) provides an attractive target in the search for anti-HIV therapies. The nonnucleoside inhibitors (NNIs) are a diverse set of compounds (usually HIV-1 specific) that function by distorting the polymerase active site upon binding in a nearby pocket. Despite being potent and of generally low toxicity, their clinical use has been limited by rapid selection for resistant viral populations. The 2.65-Å resolution structure of the complex between HIV-1 RT and the bis(heteroaryl)piperazine (BHAP) NNI, 1-(5-methanesulfonamido-1H-indol-2-yl-carbonyl)-4-[3-(1-methyl-ethylamino)pyridinyl] piperazine (U-90152), reveals the inhibitor conformation and bound water molecules. The bulky U-90152 molecule occupies the same pocket as other NNIs, but the complex is stabilized quite differently, in particular by hydrogen bonding to the main chain of Lys-103 and extensive hydrophobic contacts with Pro-236. These interactions rationalize observed resistance mutations, notably Pro-236-Leu, which occurs characteristically for BHAPs. When bound, part of U-90152 protrudes into the solvent creating a channel between Pro-236 and the polypeptide segments 225–226 and 105–106, giving the first clear evidence of the entry mode for NNIs. The structure allows prediction of binding modes for related inhibitors [(altrylamino)piperidine–BHAPs] and suggests changes to U-90152, such as the addition of a 6 amino group to the pyridine ring, which may make binding more resilient to mutations in the RT. The observation of novel hydrogen bonding to the protein main chain may provide lessons for the improvement of quite different inhibitors.
Keywords: delavirdine, AIDS, drug design, x-ray crystallography
The reverse transcriptase (RT; EC 2.7.7.49) of human immunodeficiency virus (HIV) is a multifunctional enzyme critical to the viral life cycle. It is also without a homologue in eukaryotic systems, and thus is an attractive target for anti-HIV therapies. Several RT inhibitors that act as DNA chain-terminating analogues of the natural nucleosides have been used to control HIV infection, such as AZT, ddI, ddC, d4T and 3TC (1). However, the emergence of resistant virus populations and significant adverse reactions (2, 3) led to extensive efforts to identify new RT inhibitors.
The search for novel RT inhibitors rapidly identified the nonnucleoside inhibitors (NNIs), compounds that are typically HIV-1 specific. The NNIs now comprise a very large number of chemically diverse (but largely hydrophobic) compounds, which are subdivided into groups based on their chemical structures. From early examples {e.g., 1-(2-hydroxyethoxymethyl-6-(phenylthio)thymine (HEPT) (4), tetrahydroimidazo-[4,5,l-jk][1,4]-benzodiazepin-2(1H)-one (TIBO) (5), and nevirapine (6)} to the recent example of alkenyldiarylmethanes (7), more than 30 groups of NNIs have been identified (8). These compounds can be very potent inhibitors of RT, with low toxicity and favorable pharmacokinetic properties. However, the emergence of resistant viral populations, often within days or weeks, seriously compromises their potential therapeutic efficacy. Resistance studies suggest that the NNIs share a common mode of action, binding at a single site that is distinct from the polymerase catalytic site (8). Mutations that confer resistance to one NNI often confer cross-resistance to many other inhibitors [such as the Lys-103-Asn and Tyr-181-Cys mutations (9–11)] and so interest has focused on those compounds that retain pronounced activity against mutant RTs or that have a unique resistance profile.
The first structure determination for RT [in complex with the NNI nevirapine at 3.5-Å resolution (12)] has been followed by structures of complexes with other NNIs (13–17), a complex with double-stranded DNA (18), and structures of the unliganded RT (19–21). The structures we have determined to 2.2-Å resolution for RT in complex with NNIs (13) represent the clearest models of RT to date; hence, we use the secondary structure nomenclature of these descriptions. RT is a heterodimer, one subunit (p66) comprises 560 residues and all enzymatic activity of the heterodimer is associated with residues in this subunit. The second chain (p51) comprises the initial 440 residues of the p66 subunit. RT is composed of nine domains (12), named fingers, palm, thumb, connection, and RNase H in the p66, with the p51 containing only the first four of these. The domains of the p66 subunit form a binding groove for the polynucleotide substrate, at the bottom of which lie the aspartyl residues implicated in polymerase activity.
Inhibition of RT by NNIs appears to be primarily due to distortion of the polymerase active site; a suggestion (12) subsequently supported by both structural (20) and kinetic (22) data. On binding, the NNI creates and occupies a pocket near to the active site. This process results in a switch to a stable, but inactive, conformation for the polymerase site, where the catalytic aspartyl residues are displaced relative to the binding groove as a whole (20). It appears that NNI binding also restricts the flexibility of some p66 subunit domains since several crystallization conditions require an NNI to be present (12, 23) (although the NNIs are not subsequently involved in crystal contacts) and the rate of substrate binding is higher in the presence of NNIs (22).
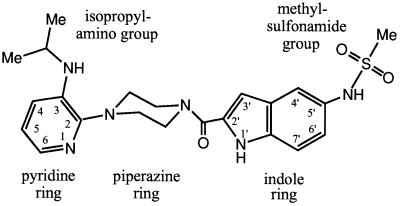
The inhibitor U-90152 (24) {1-(5-methanesulfonamido-1H-indol-2-yl-carbonyl)-4-[3-(1-methyl-ethylamino)pyridinyl] piperazine (U-90152) or delavirdine; Fig. 1} is a member of the bis(heteroaryl)piperazine (BHAP) (25) group of NNIs. BHAPs select for common resistance mutations such as Leu-100-Ile, Lys-103-Asn, and Tyr-181-Cys (10, 26), but they also select for the Pro-236-Leu mutation (27). Although the Pro-236-Leu mutation confers resistance against BHAPs, it increases the effectiveness of other NNIs (27, 28). These observations, the larger bulk of BHAPs compared with most NNIs and the use of U-90152 for clinical trials, makes the structure of the complex between RT and U-90152 a prime target for structural study.
Figure 1.
Structure of BHAP U-90152.
We report the structure of HIV-1 RT in complex with U-90152 (hereafter referred to as the “RT–BHAP complex”) refined to an R factor of 0.222 against x-ray data in the resolution range of 20–2.65 Å (Table 1). Several features of this complex are unique among the RT–NNI structures reported to date (13–17, 29), explaining the distinctive properties of BHAPs.
Table 1.
Crystallographic structure determination statistics
| Data collection details | ||
| Collection site | BL4 ESRF, 1995 | |
| Wavelength, Å | 0.995 | |
| Collimation, mm | 0.05 × 0.05 | |
| No. of crystals | 1 | |
| Unit cell dimensions, Å | 136.8 × 109.4 × 72.0 (cell form E) | |
| Data processing details | All data | Outer shell |
| Resolution range, Å | 20–2.65 | 2.74–2.65 |
| No. of observations | 83.791 | 5,304 |
| Unique reflections | 27,792 | 2,086 |
| Completeness, % | 86.8 | 69.0 |
| Reflections with F/σ(F) > 3 | 24,040 | 1,133 |
| Rmerge, %* | 10.3 | 43.9 |
| Refinement statistics | ||
| Resolution range, Å | 20–2.65 | |
| Unique reflections | 27,792 | |
| R factor, %† | 0.222 | |
| No. of protein atoms | 7765 | |
| No. of inhibitor atoms | 32 | |
| No. of water molecules | 72 | |
| rms bond length deviation, Å | 0.006 | |
| rms bond angle deviation, ° | 1.3 | |
| Mean B factors, Å2‡ | 43/50/29/74 | |
| rms B factor deviation, Å2§ | 4.7 | |
rms, root-mean-square.
Rmerge = ∑|I − <I>|/∑<I>.
R factor = ∑|Fobs − Fcalc|/∑Fobs.
Mean B factors for main chain, side chain, water, and inhibitor atoms, respectively.
rms deviation between B factors for bonded main chain atoms.
MATERIALS AND METHODS
Materials and Crystal Preparation.
The sample of U-90152 was synthesized by R. Kirsch (Hoechst, Frankfurt) and provided by J.-P. Kleim (Hoechst, Frankfurt). HIV-1 RT was expressed and purified and crystals of RT in complex with a weakly binding HEPT analogue were grown following our established procedures for obtaining crystals of other RT–NNI complexes (23). The HEPT analogue (provided by J. Chan, Glaxo Wellcome, Research Triangle Park, NC) was then soaked out of the crystals in solutions of increasing concentrations of polyethylene glycol, a method we described previously (13). The soaking procedure also increases the order in the crystals and provides a cryoprotectant (23, 30). Having produced crystals of unliganded RT, crystals of the complex between RT and almost any NNI can be obtained by adding the inhibitor to the final soak solution. For this experiment, the final resoaking solution contained 46% (wt/vol) PEG 3400 (Sigma), 0.8 mM U-90152, and ≈10% dimethyl sulfoxide buffered at pH 5.0 using a McIlvane citrate/phosphate buffer (17). Crystals were soaked for 3 days in this solution at 4°C prior to use.
Data Collection and Processing.
Diffraction data were collected from a single crystal of the RT–BHAP complex on BL4 at the European Synchrotron Radiation Facility (Grenoble, France) (Table 1). The crystal was flash-cooled to, and maintained at, 100K using an Oxford Cryosystems Cryostream (Long Hanborough, U.K.). Sixty-eight 1.5° oscillation images were recorded using 15-sec exposures on a 30-cm MAR Research imaging plate system programmed to collect in 18-cm mode and placed 212 mm from the crystal. Images were indexed and spot intensities were integrated and corrected for Lorentz and polarization effects using denzo (31), the resulting dataset being 86.8% complete for reflections in the resolution range 20–2.65 Å (Table 1). The incompleteness of the data is in a large part due to the dependence of the diffraction limit and mosaicity on crystal orientation (the mosaic spread varied from 0.4° to 1.5°). From the cell parameters we classify this crystal as being of form E in our set of related P212121 crystal forms (16, 20, 23).
Structure Determination and Refinement.
The structure of the RT–BHAP complex was solved by molecular replacement from the 2.55-Å resolution structure of the RT–MKC-442 complex (17), which had a similar cell form. The model for the protein was refined against the RT–BHAP data; first as a single rigid body, then as two rigid subunits, and finally as nine rigid domains. Positional energy minimization, simulated annealing, and restrained atomic B factor refinement followed, performed with x-plor3.1 (32) using the Engh and Huber parameter set (33). Five percent of reflections were reserved for cross-validation protocols. Due to the low reflections-to-parameters ratio all refinements used strong stereochemical restraints and the positions of atoms distant from the NNI-binding site (defined as >25 Å from the Cα atom of Tyr-188) were tightly restrained to their positions after domain-wise rigid-body refinement.
To compensate for the anisotropy and the weakness of high-resolution reflections, the dataset was sharpened using anisotropic B factor scaling against structure factors calculated from a model where the atomic B factors were scaled down by 25%. This procedure led to a dramatic improvement in the R factor for the higher resolution data. The sharpened dataset was used for further positional and restrained atomic B factor refinement incorporating a bulk solvent correction. Refinement reduced the R factor for the data from 20–2.65 Å resolution from 0.354 to 0.237, and the R factor for the cross-validation data fell from 0.362 to 0.313. Electron density maps phased from the model clearly showed the U-90152 and structural changes in the protein.
A model and suitable x-plor parameter set for U-90152 were created based on the crystal structure of BHAP U-87201 (atevirdine) deposited with the Cambridge Crystallographic Data Centre (34). A model for the complex was built into the electron density using frodo (35) on an Evans and Sutherland ESV workstation. Having optimized the refinement protocol using cross-validation, subsequent refinement used the complete x-ray dataset from 20–2.65 Å resolution to minimize overfitting of the data. Further positional and restrained atomic B factor refinements (incorporating a bulk solvent correction) along with minor rebuilding and the addition of 72 water molecules resulted in the current model (Table 1). This model has an R factor of 0.222, very good stereochemistry (typified by root-mean-square deviations of bond lengths and bond angles from their target values of 0.006 Å and 1.3°, respectively) and 88% of residues having main chain conformations in the most favored regions (and no residues in disallowed regions) of the Ramachandran plot (36). For small sections of the protein the electron density shows disorder and these parts are omitted from our model; for the p66 subunit, residues 1 and 540–560 and for the p51 subunit residues 1–5, 89–95, 216–231, and 434–440 are omitted.
Structure Analysis and Visualization.
Figs. 1 and 3b were drawn using CSC ChemDraw (Cambridge Scientific Computing, Cambridge, MA). Figs. 2, 3 a and c, and 4 were created using bobscript (R.M.E., unpublished program), a modified version of molscript (37). Figs. 3 a and c and 4 were rendered using raster3D (38, 39). The NNI pocket surface was calculated using the program volumes (R.M.E., unpublished program) to define the volume accessible to a 1.4-Å radius probe for a model of RT excluding water and inhibitor molecules but including all hydrogen atoms.
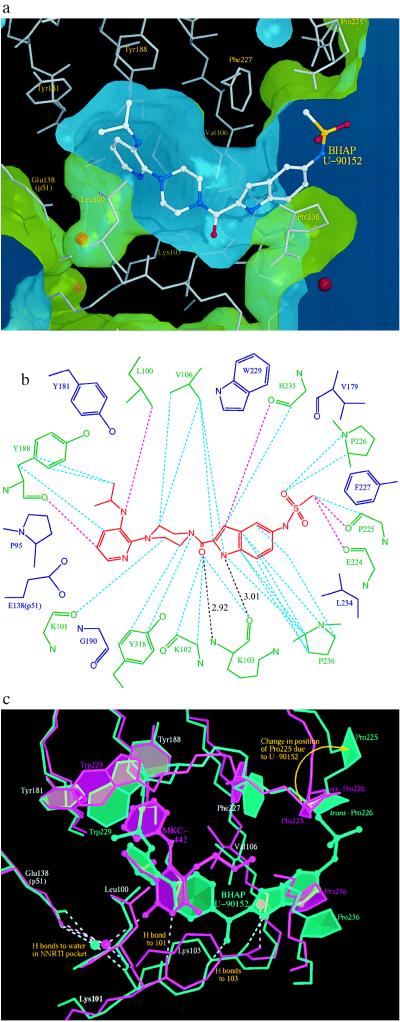
Figure 3.
Positioning of U-90152 in the NNI-binding pocket. (a) The surface of the NNI-binding pocket in the complex with U-90152. The inhibitor is shown as an atom-colored ball-and-stick model with the surrounding protein structure as thin grey sticks. Water molecules are indicated by red spheres. The green face of the surface points toward the protein; the blue face toward the solvent. This is the first RT–NNI complex structure to show the channel connecting the pocket to the solvent and may indicate the mode of entry into the (normally buried) pocket for all NNIs. (b) A diagram of the contacts between U-90152 (in red) and the surrounding protein structure. Specific interactions are shown by broken lines (pink, <3.3 Å; blue, 3.3–3.6 Å; black, hydrogen bonds with distances). Residues contacting U-90152 are shown in green, those observed in contact with NNIs in other structures are in blue. (c) A comparison between the RT complexes with the NNIs U-90152 (green) and MKC-442 (17) (pink). The inhibitor is shown as a ball-and-stick representation, the surrounding protein structure as sticks, water molecules as spheres, and hydrogen bonds as broken lines. To compensate for the (slight) difference in crystal forms the models were superposed based on the surrounding protein structure in the manner we have described (13). Only the pyridine end of U-90152 occupies equivalent volume to MKC-442 (and indeed all other NNIs studied to date). The novel hydrogen bonding pattern and significant effects on Val-106, Pro-225, and Pro-236 are clearly shown.
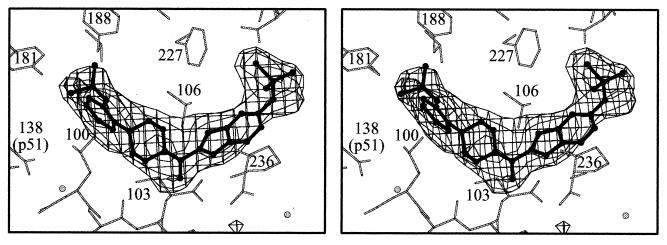
Figure 2.
Stereo diagram showing Fobs − Fcalc omit electron density for U-90152 contoured at 3σ U-90152 is shown in ball-and-stick representation and the surrounding protein structure is shown by thin sticks. Residue Tyr-318, which would otherwise obscure the BHAP carbonyl group, is omitted from the figure for clarity.
RESULTS
Structure of the RT Heterodimer.
Except for localized differences arising from the binding of U-90152 (which are described below), our general description of the protein structure for the RT–nevirapine complex (13) also applies to the RT–BHAP complex.
Positioning of U-90152 in the NNI Pocket.
Despite the larger bulk of U-90152 compared with other NNIs we have studied [volume of inhibitors: U-90152, 380 Å3; TNK-651, 320 Å3; all others, 230–290 Å3 (13, 16, 17)], electron density for the inhibitor (Fig. 2) clearly shows that it binds to RT in the NNI-binding pocket. Its size and shape cause the molecule to extend beyond the usual pocket and to project into the solvent (Fig. 3a). On binding other NNIs, the side chains of residues surrounding the inhibitor (especially Tyr-181 and Tyr-188) rearrange and the nearby main chain (especially strands β4, β7, and β8) moves to form the pocket (20). The RT–BHAP structure shows essentially the same inhibited conformation for the protein, with the catalytic aspartyl residues moved relative to the unliganded conformation (20).
Fig. 3b shows the interactions between U-90152 and the residues lining the binding pocket. The most buried part of U-90152 is the (isopropylamino)pyridino group. The isopropylamino group is similarly positioned to the dimethylallyl group in our RT–Cl-TIBO complex (16) and it plays an equivalent role, interacting with the rings of Tyr-188 and Trp-229 (minimum distance between U-90152 and Trp-229 is 3.8 Å). The pyridine ring is positioned near the rearranged tyrosyl residues 181 and 188 (distance from C5 of the pyridine ring to both Cβ atoms is 3.7 Å) and it occupies part of the volume that these side chains occupy in the unliganded structure (20). As U-90152 enters the pocket it is presumably this ring that triggers the rearrangement (17). Moving along U-90152, the piperazine ring conformation brings the inhibitor very close to Val-106 and into contact with a set of residues different from those contacted by other NNIs. Two of these novel interactions are particularly important for stabilizing the RT–BHAP complex; hydrogen bonding to the main chain of Lys-103 and strong hydrophobic interactions between the indole ring of U-90152 and Pro-236.
Hydrogen Bonding by U-90152.
Not all NNIs make direct hydrogen bonds with RT [e.g., nevirapine and its analogues (13, 29)], but many NNIs, including HEPT and TIBO analogues (13, 15–17), form a hydrogen bond to the carbonyl oxygen of Lys-101 (Fig. 3c). U-90152 makes instead a novel pair of hydrogen bonds to the main chain of Lys-103 via its carbonyl oxygen and indole nitrogen atoms (Fig. 3 b and c). The bulk of U-90152 and the presence of these hydrogen bonds affects the nearby protein structure. The position and conformation of residues 101–103 are similar to those seen in the complex with another bulky NNI, TNK-651 (17). Whereas in that case a bend at residue 104 brings the main chain back into register with other structures by residue 106 (17), for the RT–BHAP complex, the bend is at residue 105 and the structures only come into register at residue 108 (Fig. 3c). The result is a 0.9 Å shift in the position of the Cβ atom of Val-106, which brings this residue into close contact with U-90152. The observation of a new hydrogen bonding pattern suggests that it may be possible to design molecules that interact with the main chain atoms of residues 101 and 103, thus creating inhibitors that are less susceptible to escape by a point mutation.
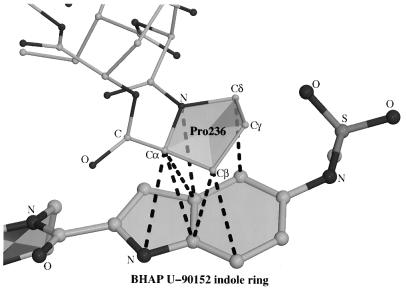
Interaction Between Indole Ring and Pro-236.
Interactions between aromatic rings stabilize many RT–NNI complexes (13–17, 29). With U-90152, a nonaromatic ring interaction between the inhibitor indole ring and Pro-236 is particularly important (Fig. 4), with seven inter-ring atom distances being less than 3.6 Å (Fig. 3b). In RT–NNI complexes, Pro-236 has been seen to adopt a wide “fan” of conformations (17), all corresponding to more compact structures than observed with the unliganded enzyme (20). In the RT–BHAP complex structure however, the presence of the indole ring forces Pro-236 into an even more open conformation, partly accounting for the connecting channel between the NNI-binding pocket and the bulk solvent.
Figure 4.
Interactions between the indole ring of U-90152 and Pro-236. U-90152 is shown with thick bonds, residues 235–237 with thin bonds and interatomic distances <3.6 Å by broken lines. With so many interactions it is not surprising that mutations of this residue (such as Pro-236-Leu) disrupt the binding of BHAPs.
Bound Water in the NNI Pocket.
In common with the water structure we observe in other RT–NNI complexes (13, 16, 17), there is a water molecule in the NNI pocket forming hydrogen bonds to the main chain nitrogen of Lys-101 and the side chain of Glu-138 from the p51 subunit (Fig. 3 a and c). Unlike many complexes however, the U-90152 does not approach this water molecule closely enough to make a hydrogen bond. Elsewhere, the indole nitrogen of U-90152 is similarly positioned to a water molecule observed in the RT–MKC-442 complex (17) and this water molecule makes an equivalent hydrogen bond to Lys-103 (Fig. 3c).
Flexibility of the RT–BHAP Complex.
While the electron density shows the U-90152 clearly, the atomic B factors for the inhibitor and surrounding protein suggest that only the buried parts of the inhibitor contribute to the rigidity of the complex. Parts of the indole ring and particularly the methylsulfonamide group have very high B factors and there is evidence for a second conformation of the latter group. The atomic B factors for residues 223–226 are similarly high. Such mobility reflects binding to a flexible part of the protein, but it also suggests that replacing the methylsulfonamide group by a smaller group may be preferable. In its predominant conformation, the methylsulfonamide group distorts the conformation of residues 223–226 by displacing Pro-225, which swings around to open up the entrance to the NNI pocket (Fig. 3c). In this process, the cis-proline at residue 226 (a conformation we have observed in all liganded and unliganded structures to date) is switched into the trans conformation.
Entry into the NNI-Binding Pocket.
Speculation as to how NNIs enter the binding pocket has flourished in the absence of firm evidence (12–14). Due to the size of U-90152, the inhibitor projects out of the pocket and into the bulk solvent (Fig. 3a). The pocket is thus connected to the solvent by a channel between a well-ordered sheet (strands β4, β7, and β8) and a highly mobile flap (strands β9, β10, and β11), strongly suggesting that entry of U-90152 is by means of this channel. This is the first time that such a connection has been observed and it may have implications for understanding the binding process for all NNIs. Assuming the same mode of entry for smaller NNIs, the strands β9, β10, and β11 act as a “trap door,” opening to let the inhibitor in and closing behind it once it has entered.
DISCUSSION
Pro-236-Leu Mutation.
One of the intriguing features of BHAPs is the spectrum of resistance mutations for which they select (Table 2). Most interesting among these mutations is the Pro-236-Leu mutation that is observed for BHAPs and that increases sensitivity of RT to other NNIs (27, 28). Pro-236 is the residue making the most extensive contacts with U-90152 (Fig. 4). Thus, a mutation that disrupts these contacts might be expected to be an effective resistance mutation. Mutagenesis experiments have measured the degree of resistance conferred by mutations at residue 236 (Pro-236-Leu, 25-fold; Pro-236-Thr, 8-fold; Pro-236-His, 6-fold; Pro-236-Arg, 3-fold; Pro-236-Ala, 2-fold) and shown that these correlate with sensitization to the NNI L-697,661 (28). The suggestion on this evidence that Pro-236 does not contact BHAPs directly (28) is clearly not consistent with the data reported here. We observe that effective resistance is conferred by residues with branched side chains, since branching may hinder the formation of strong interactions with the planar BHAP indole ring. The structure suggests that replacement of the indole ring by a pyrrole ring (40) may be worth reevaluating since such a change may reduce the degree of resistance due to the Pro-236-Leu mutation.
Table 2.
Relative resistance to U-90152 conferred by different RT mutations
| L100I | K103N | V106A | V179A | Y181C | P236L | |
|---|---|---|---|---|---|---|
| Dueweke et al. (24)* | — | 30× | — | — | 32× | — |
| Dueweke et al. (27)* | — | 30× | — | — | 32× | 70× |
| Romero et al. (40)* | — | — | — | — | 8× | >100× |
| Fan et al. (28)† | — | — | — | — | — | 25× |
| Balzarini et al. (41)‡ | 77× | 45× | 27× | 22× | 42× | — |
—, Not determined.
Ratio of in vitro IC50 values.
Estimated ratio of IC50 values.
Ratio of EC50 values.
Other Mutations.
Other mutations tend to correspond to changes seen in response to other NNIs and may be similarly rationalized (13, 16).
Leu-100-Ile mutation.
An isoleucyl residue cannot be accommodated without substantial structural change, because in many plausible conformations the Cγ2 atom of Ile-100 clashes badly with one of the carbon atoms in the U-90152 piperazine ring (even in the “best” orientation the atomic separation is only 2.9 Å). RT bearing the Leu-100-Ile mutation remains sensitive to a closely related group of NNIs, the (alkylamino)piperidine bis(heteroaryl)piperazine analogues (AAP–BHAPs) (42), in which an extra nitrogen atom is inserted between the piperidine ring (replacing the BHAP piperazine ring) and the pyridine ring. Modelling the binding of AAP–BHAPs based on the RT–BHAP structure suggests that repositioning of these rings due to the insertion may account for the continued sensitivity.
Lys-103-Asn mutation.
Although lysine is a charged residue, in the context of the NNI-binding pocket it acts as a hydrophobic residue forming part of the base of the pocket with its charged tip exposed to the solvent. Mutation to a shorter polar residue would alter the properties of the pocket and reduce the net gain in hydrophobic interactions on moving from unliganded to NNI-bound RT. There may also be a more specific component to loss of potency since all observed changes to this residue are to hydrogen bond acceptors (J. Balzarini, personal communication). With BHAP, the pyridine may form a hydrogen bond to the aspariginyl residue in the mutant RT preventing binding in the correct mode.
Val-106-Ala mutation.
In the complex, the central part of U-90152 closely contacts the Cγ atoms of Val-106, which thus, along with Lys-103, help to dictate the conformation of the bound inhibitor (Fig. 3c). The mode of escape by this mutation is therefore likely to be similar to Lys-103 mutations and so it is unsurprising that the Lys-103-Asn/Val-106-Ala double mutation is particularly effective against a variety of NNIs (J. Balzarini, H. Pelemans and E. deClercq, unpublished work).
Interaction Between BHAP and RT.
The key factor leading to the unusual spectrum of RT resistance mutations against BHAPs is the greater size of BHAPs, which allows interaction with parts of the NNI-binding pocket inaccessible to other NNIs. Two major interactions stabilizing the RT–BHAP complex have not been observed before; hydrogen bonds to Lys-103 and extensive hydrophobic interactions with Pro-236. It is interesting to note that the BHAP carbonyl and NH groups that form hydrogen bonds to Lys-103 are reminiscent of an amide group, strengthening the structural analogy between the NNIs and the polypeptide chain that fills the pocket in the p51 subunit (20).
During optimization of the BHAPs, it was found that compounds not containing an indole ring behaved differently, for instance with aryl rings the neighboring carbonyl group can be replaced by a methylene linkage with little effect on potency. The opposite behavior was observed for changes to the pyridine nitrogen, which is crucial for activity with aryl-containing compounds (40). These observations suggest that BHAPs without indole rings (including the lead compound) may have a different binding mode from U-90152, perhaps with the heterocycle positions exchanged so that the pyridine nitrogen interacts with residue 103.
We have previously noted the importance of NNIs having atoms positioned to perturb the conformation of Tyr-181 (13, 16, 17). From this perspective, the addition of a 6-position amino group to the BHAP pyridine ring may be favorable and could make interactions with the main chain oxygen of Val-179. It is worth noting that AAP–BHAPs retain significant inhibitory activity against many mutant RTs, such as the Leu-100-Ile mutation discussed above and the Tyr-181-Cys mutation [which only confers 7- to 10-fold resistance to the AAP–BHAPs (42)]. It may be that in complexes with AAP–BHAPs the pyridine ring is better positioned to perturb Tyr-181 than the equivalent ring with U-90152.
Finally, the novel hydrogen bonding pattern observed between the protein and the NNI U-90152 may be quite a general vehicle for generating binding affinity that is less susceptible to point mutations. It seems likely that such interactions could be engineered into other inhibitor complexes, perhaps with quite different chemistry.
Acknowledgments
We thank Drs. R. Kirsch and J.-P. Kleim for their kind gift of U-90152, Dr. J. Chan for the gift of the HEPT analogue, Dr. B. Rasmussen (European Molecular Biology Laboratory) and Dr. E. Mitchell (European Synchrotron Radiation Facility) for their help with x-ray data collection, and Prof. J. Balzarini (Rega Institute, Leuven) for helpful discussions about NNI resistance. We would also like to thank Dr. R. Bryan for computing support and Mr. S. Lee for help with the preparation of figures. R.M.E. is now supported by the Belgian Nationaal Fonds voor Wetenschappelijk Onderzoek (Grant G. 3304.96) and E.Y.J. is a Royal Society University Research Fellow. The Oxford Centre for Molecular Sciences is supported by the Biotechnology and Biological Sciences Research Council and the Medical Research Council of Great Britain and this project has received the long-term support of the Medical Research Council AIDS-Directed Programme.
ABBREVIATIONS
- RT
reverse transcriptase
- NNI
nonnucleoside reverse transcriptase inhibitor
- BHAP
bis(heteroaryl)piperazine
- U-90152
1-(5-methanesulfonamido-1H-indol-2-yl-carbonyl)-4-[3-(1-methyl-ethylamino)pyridinyl] piperazine
- RT–BHAP complex
complex between RT and BHAP U-90152
- AAP–BHAP
(alkylamino)piperidine bis(heteroaryl)piperazine analogue
Footnotes
Data deposition: The atomic coordinates for the RT–BHAP complex have been deposited with the Protein Data Bank, Chemistry Department, Brookhaven National Laboratory, Upton, NY 11973 (references 1KLM and R1KLMSF for structure factors), for release 1 year from the date of publication.
References
- 1.De Clercq E. J Med Chem. 1995;38:2491–2517. doi: 10.1021/jm00014a001. [DOI] [PubMed] [Google Scholar]
- 2.Richman D D, Fischl M A, Grieco M H, Gottleib M S, Volberding P A, Laskin O L, Leedom J M, Groopman J E, Mildvan D, Hirsch M S, Jackson G G, Durack D T, Nusinoff-Lehrman S the AZT Collaborative Working Group. N Engl J Med. 1987;317:192–197. doi: 10.1056/NEJM198707233170402. [DOI] [PubMed] [Google Scholar]
- 3.Larder B A, Kemp S D. Science. 1989;246:1155–1158. doi: 10.1126/science.2479983. [DOI] [PubMed] [Google Scholar]
- 4.Baba M, Tanaka H, De Clercq E, Pauwels R, Balzarini J, Schols D, Nakashima H, Perno C-F, Walker R T, Miyasaka T. Biochem Biophys Res Commun. 1989;165:1375–1381. doi: 10.1016/0006-291x(89)92756-3. [DOI] [PubMed] [Google Scholar]
- 5.Pauwels R, Andries K, Desmyter J, Schols D, Kukla M J, Breslin H J, Raeymaeckers A, Gelder J V, Woestenborghs R, Heykants J, Schellekens K, Janssen M A C, De Clercq E, Janssen P A J. Nature (London) 1990;343:470–474. doi: 10.1038/343470a0. [DOI] [PubMed] [Google Scholar]
- 6.Merluzzi V J, Hargrave K D, Labadia M, Grozinger K, Skoog M, Wu J C, Shih C-K, Eckner K, Hattox S, Adams J, Rosenthal A S, Faanes R, Eckner R J, Koup R A, Sullivan J L. Science. 1990;250:1411–1413. doi: 10.1126/science.1701568. [DOI] [PubMed] [Google Scholar]
- 7.Cushman M, Golebiewski W M, Graham L, Turpin J A, Rice W G, Fliakas-Boltz V, Buckheit R W., Jr J Med Chem. 1996;39:3217–3227. doi: 10.1021/jm960082v. [DOI] [PubMed] [Google Scholar]
- 8.De Clercq E. Rev Med Virol. 1996;6:97–117. doi: 10.1002/(SICI)1099-1654(199606)6:2<97::AID-RMV168>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 9.Nunberg J H, Schleif W A, Boots E J, O’Brien J A, Quintero J C, Hoffman J M, Jr, Emini E A, Goldman M E. J Virol. 1991;65:4887–4892. doi: 10.1128/jvi.65.9.4887-4892.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Balzarini J, Karlsson A, Pérez-Pérez M-J, Camarasa M-J, Tarpley W G, De Clercq E. J Virol. 1993;67:5353–5359. doi: 10.1128/jvi.67.9.5353-5359.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Balzarini J, Karlsson A, Pérez-Pérez M-J, Vrang L, Walbers J, Zhang H, Öberg B, Vandamme A-M, Camarasa M-J, De Clercq E. Virology. 1993;192:246–253. doi: 10.1006/viro.1993.1027. [DOI] [PubMed] [Google Scholar]
- 12.Kohlstaedt L A, Wang J, Friedman J M, Rice P A, Steitz T A. Science. 1992;256:1783–1790. doi: 10.1126/science.1377403. [DOI] [PubMed] [Google Scholar]
- 13.Ren J, Esnouf R, Garman E, Somers D, Ross C, Kirby I, Keeling J, Darby G, Jones Y, Stuart D, Stammers D. Nat Struct Biol. 1995;2:293–302. doi: 10.1038/nsb0495-293. [DOI] [PubMed] [Google Scholar]
- 14.Ding J, Das K, Tantillo C, Zhang W, Clark A D, Jr, Jessen S, Lu X, Hsiou Y, Jacobo-Molina A, Andries K, Pauwels R, Moereels H, Koymans L, Janssen P A J, Smith R H J, Kroege Koepke R, Michejda C J, Hughes S H, Arnold E. Structure (London) 1995;3:365–379. doi: 10.1016/s0969-2126(01)00168-x. [DOI] [PubMed] [Google Scholar]
- 15.Ding J, Das K, Moereels H, Koymans L, Andries K, Janssen P A J, Hughes S H, Arnold E. Nat Struct Biol. 1995;2:407–415. doi: 10.1038/nsb0595-407. [DOI] [PubMed] [Google Scholar]
- 16.Ren J, Esnouf R, Hopkins A, Ross C, Jones Y, Stammers D, Stuart D. Structure (London) 1995;3:915–926. doi: 10.1016/S0969-2126(01)00226-X. [DOI] [PubMed] [Google Scholar]
- 17.Hopkins A L, Ren J, Esnouf R M, Willcox B E, Jones E Y, Ross C, Miyasaka T, Walker R T, Tanaka H, Stammers D K, Stuart D I. J Med Chem. 1996;39:1589–1600. doi: 10.1021/jm960056x. [DOI] [PubMed] [Google Scholar]
- 18.Jacobo-Molina A, Ding J, Nanni R G, Clark A D, Jr, Lu X, Tantillo C, Williams R L, Kamer G, Ferris A L, Clark P, Hizi A, Hughes S H, Arnold E. Proc Natl Acad Sci USA. 1993;90:6320–6324. doi: 10.1073/pnas.90.13.6320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rodgers D W, Gamblin S J, Harris B A, Ray S, Culp J S, Hellmig B, Woolf D J, Debouck C, Harrison S C. Proc Natl Acad Sci USA. 1995;92:1222–1226. doi: 10.1073/pnas.92.4.1222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Esnouf R, Ren J, Ross C, Jones Y, Stammers D, Stuart D. Nat Struct Biol. 1995;2:303–308. doi: 10.1038/nsb0495-303. [DOI] [PubMed] [Google Scholar]
- 21.Hsiou Y, Ding J, Das K, Clark A D, Jr, Hughes S H, Arnold E. Structure (London) 1996;4:853–860. doi: 10.1016/s0969-2126(96)00091-3. [DOI] [PubMed] [Google Scholar]
- 22.Spence R A, Kati W M, Anderson K S, Johnson K A. Science. 1995;267:988–993. doi: 10.1126/science.7532321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stammers D K, Somers D O, Ross C K, Kirby I, Ray P H, Wilson J E, Norman M, Ren J S, Esnouf R M, Garman E F, Jones E Y, Stuart D I. J Mol Biol. 1994;242:586–588. doi: 10.1006/jmbi.1994.1604. [DOI] [PubMed] [Google Scholar]
- 24.Dueweke T J, Poppe S M, Romero D L, Swaney S M, So A G, Downey K M, Althaus I W, Reusser F, Busso M, Resnick L, Mayers D L, Lane J, Aristoff P A, Thomas R C, Tarpley W G. Antimicrob Agents Chemother. 1993;37:1127–1131. doi: 10.1128/aac.37.5.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Romero D L, Busso M, Tan C-K, Reusser F, Palmer J R, Poppe S M, Aristoff P A, Downey K M, So A G, Resnick L, Tarpley W G. Proc Natl Acad Sci USA. 1991;88:8806–8810. doi: 10.1073/pnas.88.19.8806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Vasudevachari M B, Battista C, Lane H C, Psallidopoulos M C, Zhao B, Cook J, Palmer J R, Romero D L, Tarpley W G, Salzman N P. Virology. 1992;190:269–277. doi: 10.1016/0042-6822(92)91213-e. [DOI] [PubMed] [Google Scholar]
- 27.Dueweke T J, Pushkarskaya T, Poppe S M, Swaney S M, Zhao J Q, Chen I S Y, Stevenson M, Tarpley W G. Proc Natl Acad Sci USA. 1993;90:4713–4717. doi: 10.1073/pnas.90.10.4713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fan N, Evans D B, Rank K B, Thomas R C, Tarpley W G, Sharma S K. FEBS Lett. 1995;359:233–238. doi: 10.1016/0014-5793(95)00051-a. [DOI] [PubMed] [Google Scholar]
- 29.Smerdon S J, Jager J, Wang J, Kohlstaedt L A, Chirino A J, Friedman J M, Rice P A, Steitz T A. Proc Natl Acad Sci USA. 1994;91:3911–3915. doi: 10.1073/pnas.91.9.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Esnouf R, Ren J, Jones Y, Stammers D, Stuart D. In: Macromolecular Refinement. Dodson E, Moore M, Ralph A, Bailey S, editors. Warrington, U.K.: CCLRC Daresbury Lab.; 1996. pp. 153–161. [Google Scholar]
- 31.Otwinowski Z. In: Data Collection and Processing. Sawyer L, Isaacs N, Bailey S, editors. Warrington, U.K.: SERC Daresbury Lab.; 1993. pp. 56–62. [Google Scholar]
- 32.Brünger A T. x-plor. New Haven, CT: Yale Univ. Press; 1992. Version 3.1. [Google Scholar]
- 33.Engh R A, Huber R. Acta Crystallogr A. 1991;47:392–400. [Google Scholar]
- 34.Genin M J, Chidester C G, Rohrer D C, Romero D L. Bioorg Med Chem Lett. 1995;5:1875–1880. [Google Scholar]
- 35.Jones T A. Methods Enzymol. 1985;115:157–171. doi: 10.1016/0076-6879(85)15014-7. [DOI] [PubMed] [Google Scholar]
- 36.Laskowski R A, MacArthur M W, Moss D S, Thornton J M. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- 37.Kraulis P J. J Appl Crystallogr. 1991;24:946–950. [Google Scholar]
- 38.Bacon D J, Anderson W F. J Mol Graph. 1988;6:219–220. [Google Scholar]
- 39.Merritt E A, Murphy M E P. Acta Crystallogr D. 1994;50:869–873. doi: 10.1107/S0907444994006396. [DOI] [PubMed] [Google Scholar]
- 40.Romero D L, Morge R A, Genin M J, Biles C, Busso M, Resnick L, Althaus I W, Reusser F, Thomas R C, Tarpley W G. J Med Chem. 1993;36:1505–1508. doi: 10.1021/jm00062a027. [DOI] [PubMed] [Google Scholar]
- 41.Balzarini J, Pelemans H, Aquaro S, Perno C-F, Witvrouw M, Schols D, De Clercq E, Karlsson A. Mol Pharmacol. 1996;50:394–401. [PubMed] [Google Scholar]
- 42.Olmsted R A, Slade D E, Kopta L A, Poppe S M, Poel T J, Newport S W, Rank K B, Biles C, Morge R A, Dueweke T J, Yagi Y, Romero D L, Thomas R C, Sharma S K, Tarpley W G. J Virol. 1996;70:3698–3705. doi: 10.1128/jvi.70.6.3698-3705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]