Abstract
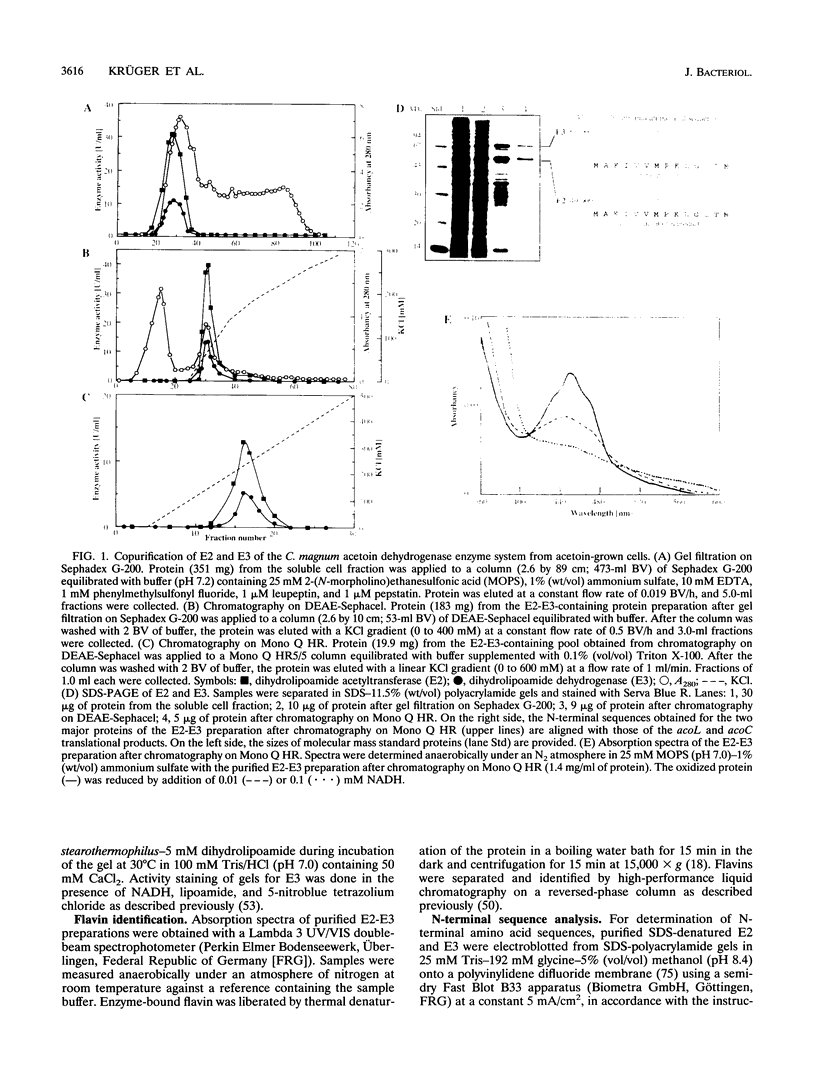
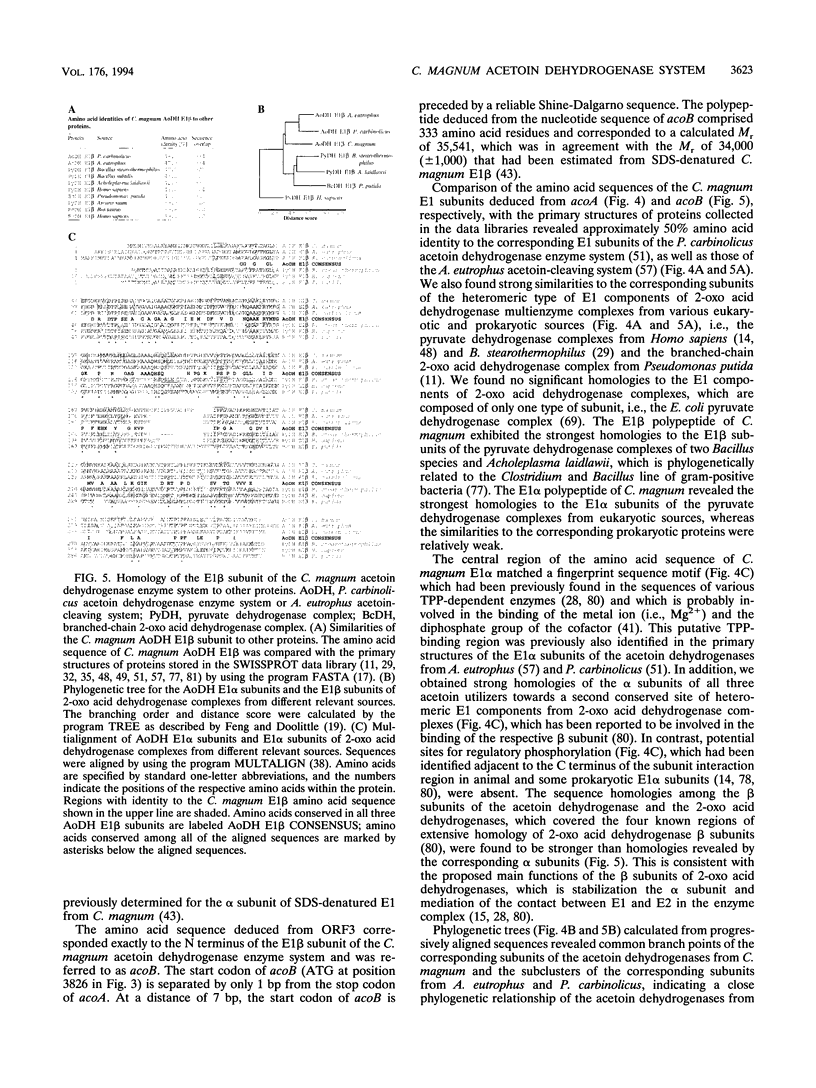
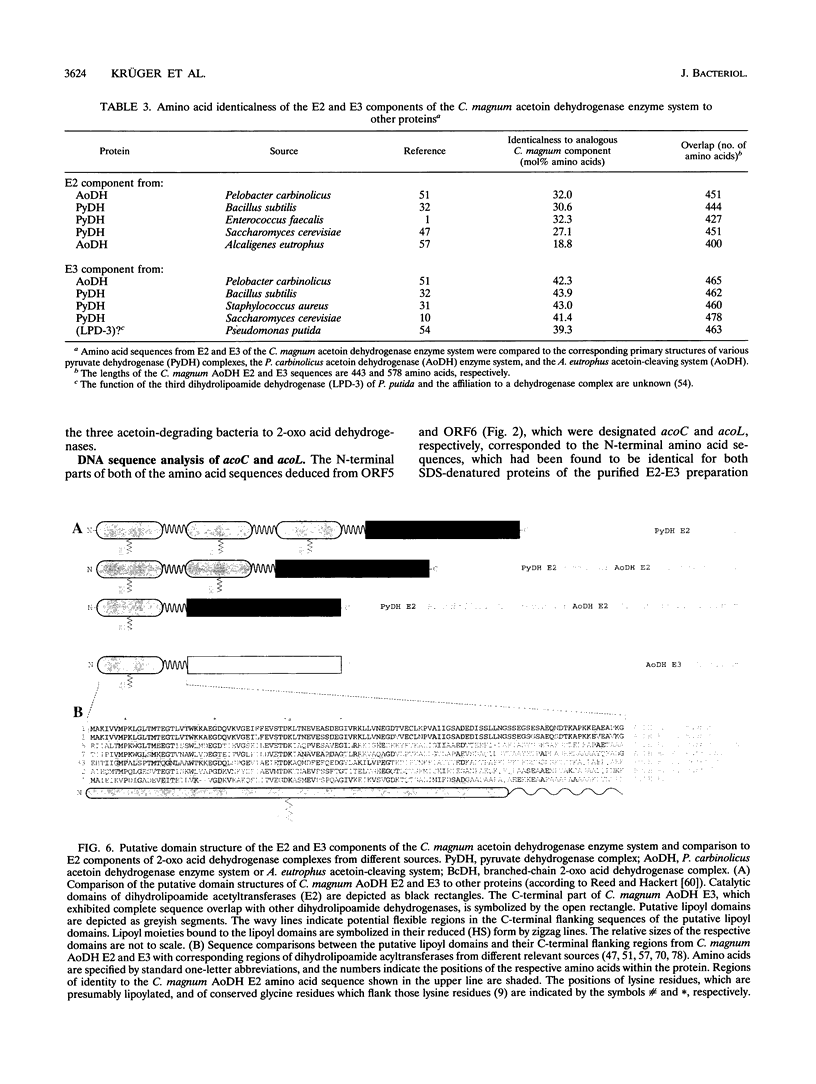
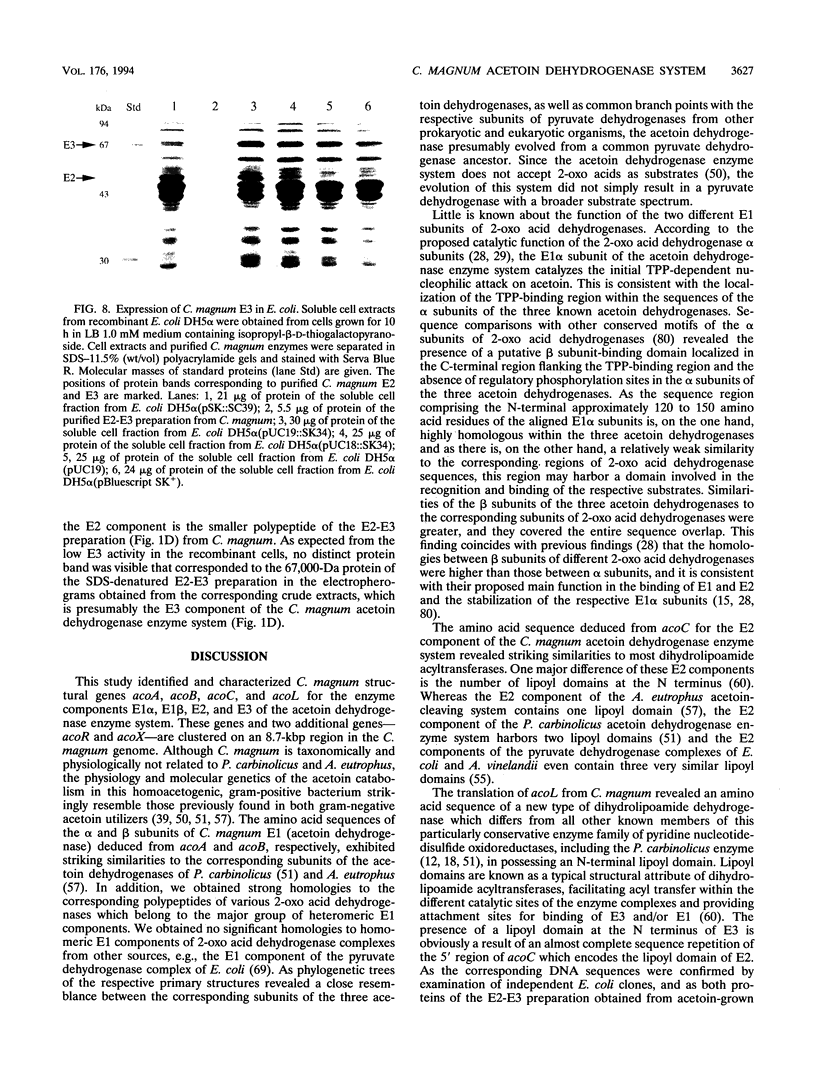
E2 (dihydrolipoamide acetyltransferase) and E3 (dihydrolipoamide dehydrogenase) of the Clostridium magnum acetoin dehydrogenase enzyme system were copurified in a three-step procedure from acetoin-grown cells. The denatured E2-E3 preparation comprised two polypeptides with M(r)s of 49,000 and 67,000, respectively. Microsequencing of both proteins revealed identical amino acid sequences. By use of oligonucleotide probes based on the N-terminal sequences of the alpha and beta subunits of E1 (acetoin dehydrogenase, thymine PPi dependent), which were purified recently (H. Lorenzl, F.B. Oppermann, B. Schmidt, and A. Steinbüchel, Antonie van Leeuwenhoek 63:219-225, 1993), and of E2-E3, structural genes acoA (encoding E1 alpha), acoB (encoding E1 beta), acoC (encoding E2), and acoL (encoding E3) were identified on a single ClaI restriction fragment and expressed in Escherichia coli. The nucleotide sequences of acoA (978 bp), acoB (999 bp), acoC (1,332 bp), and acoL (1,734 bp), as well as those of acoX (996 bp) and acoR (1,956 bp), were determined. The amino acid sequences deduced from acoA, acoB, acoC, and acoL for E1 alpha (M(r), 35,532), E1 beta (M(r), 35,541), E2 (M(r), 48,149), and E3 (M(r), 61,255) exhibited striking similarities to the amino acid sequences of the corresponding components of the Pelobacter carbinolicus acetoin dehydrogenase enzyme system and the Alcaligenes eutrophus acetoin-cleaving system, respectively. Significant homologies to the enzyme components of various 2-oxo acid dehydrogenase complexes were also found, indicating a close relationship between the two enzyme systems. As a result of the partial repetition of the 5' coding region of acoC into the corresponding part of acoL, the E3 component of the C. magnum acetoin dehydrogenase enzyme system contains an N-terminal lipoyl domain, which is unique among dihydrolipoamide dehydrogenases. We found strong similarities between the AcoR and AcoX sequences and the A. eutrophus acoR gene product, which is a regulatory protein required for expression of the A. eutrophus aco genes, and the A. eutrophus acoX gene product, which has an unknown function, respectively. The aco genes of C. magnum are probably organized in one single operon (acoABXCL); acoR maps upstream of this operon.
Full text
PDF












Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Allen A. G., Perham R. N. Two lipoyl domains in the dihydrolipoamide acetyltransferase chain of the pyruvate dehydrogenase multienzyme complex of Streptococcus faecalis. FEBS Lett. 1991 Aug 5;287(1-2):206–210. doi: 10.1016/0014-5793(91)80052-5. [DOI] [PubMed] [Google Scholar]
- Andersson L. -O., Borg H., Mikaelsson M. Molecular weight estimations of proteins by electrophoresis in polyacrylamide gels of graded porosity. FEBS Lett. 1972 Feb 1;20(2):199–202. doi: 10.1016/0014-5793(72)80793-2. [DOI] [PubMed] [Google Scholar]
- Arnold W., Rump A., Klipp W., Priefer U. B., Pühler A. Nucleotide sequence of a 24,206-base-pair DNA fragment carrying the entire nitrogen fixation gene cluster of Klebsiella pneumoniae. J Mol Biol. 1988 Oct 5;203(3):715–738. doi: 10.1016/0022-2836(88)90205-7. [DOI] [PubMed] [Google Scholar]
- Atkinson T., Hammond P. M., Hartwell R. D., Hughes P., Scawen M. D., Sherwood R. F., Small D. A., Bruton C. J., Harvey M. J., Lowe C. R. Triazine-dye affinity; chromatography. Biochem Soc Trans. 1981 Aug;9(4):290–293. doi: 10.1042/bst0090290. [DOI] [PubMed] [Google Scholar]
- Behal R. H., Browning K. S., Reed L. J. Nucleotide and deduced amino acid sequence of the alpha subunit of yeast pyruvate dehydrogenase. Biochem Biophys Res Commun. 1989 Oct 31;164(2):941–946. doi: 10.1016/0006-291x(89)91549-0. [DOI] [PubMed] [Google Scholar]
- Birnboim H. C., Doly J. A rapid alkaline extraction procedure for screening recombinant plasmid DNA. Nucleic Acids Res. 1979 Nov 24;7(6):1513–1523. doi: 10.1093/nar/7.6.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borges A., Hawkins C. F., Packman L. C., Perham R. N. Cloning and sequence analysis of the genes encoding the dihydrolipoamide acetyltransferase and dihydrolipoamide dehydrogenase components of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. Eur J Biochem. 1990 Nov 26;194(1):95–102. doi: 10.1111/j.1432-1033.1990.tb19432.x. [DOI] [PubMed] [Google Scholar]
- Browner M. F., Taroni F., Sztul E., Rosenberg L. E. Sequence analysis, biogenesis, and mitochondrial import of the alpha-subunit of rat liver propionyl-CoA carboxylase. J Biol Chem. 1989 Jul 25;264(21):12680–12685. [PubMed] [Google Scholar]
- Browning K. S., Uhlinger D. J., Reed L. J. Nucleotide sequence for yeast dihydrolipoamide dehydrogenase. Proc Natl Acad Sci U S A. 1988 Mar;85(6):1831–1834. doi: 10.1073/pnas.85.6.1831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns G., Brown T., Hatter K., Idriss J. M., Sokatch J. R. Similarity of the E1 subunits of branched-chain-oxoacid dehydrogenase from Pseudomonas putida to the corresponding subunits of mammalian branched-chain-oxoacid and pyruvate dehydrogenases. Eur J Biochem. 1988 Sep 15;176(2):311–317. doi: 10.1111/j.1432-1033.1988.tb14283.x. [DOI] [PubMed] [Google Scholar]
- Carothers D. J., Pons G., Patel M. S. Dihydrolipoamide dehydrogenase: functional similarities and divergent evolution of the pyridine nucleotide-disulfide oxidoreductases. Arch Biochem Biophys. 1989 Feb 1;268(2):409–425. doi: 10.1016/0003-9861(89)90309-3. [DOI] [PubMed] [Google Scholar]
- Chopra A. K., Peterson J. W., Prasad R. Nucleotide sequence analysis of purH and purD genes from Salmonella typhimurium. Biochim Biophys Acta. 1991 Nov 11;1090(3):351–354. doi: 10.1016/0167-4781(91)90202-w. [DOI] [PubMed] [Google Scholar]
- Dahl H. H., Brown R. M., Hutchison W. M., Maragos C., Brown G. K. A testis-specific form of the human pyruvate dehydrogenase E1 alpha subunit is coded for by an intronless gene on chromosome 4. Genomics. 1990 Oct;8(2):225–232. doi: 10.1016/0888-7543(90)90275-y. [DOI] [PubMed] [Google Scholar]
- Davie J. R., Wynn R. M., Cox R. P., Chuang D. T. Expression and assembly of a functional E1 component (alpha 2 beta 2) of mammalian branched-chain alpha-ketoacid dehydrogenase complex in Escherichia coli. J Biol Chem. 1992 Aug 15;267(23):16601–16606. [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dietrichs D., Andreesen J. R. Purification and comparative studies of dihydrolipoamide dehydrogenases from the anaerobic, glycine-utilizing bacteria Peptostreptococcus glycinophilus, Clostridium cylindrosporum, and Clostridium sporogenes. J Bacteriol. 1990 Jan;172(1):243–251. doi: 10.1128/jb.172.1.243-251.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D. F., Doolittle R. F. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25(4):351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Fitzgerald J., Hutchison W. M., Dahl H. H. Isolation and characterisation of the mouse pyruvate dehydrogenase E1 alpha genes. Biochim Biophys Acta. 1992 May 7;1131(1):83–90. doi: 10.1016/0167-4781(92)90102-6. [DOI] [PubMed] [Google Scholar]
- Flinta C., Persson B., Jörnvall H., von Heijne G. Sequence determinants of cytosolic N-terminal protein processing. Eur J Biochem. 1986 Jan 2;154(1):193–196. doi: 10.1111/j.1432-1033.1986.tb09378.x. [DOI] [PubMed] [Google Scholar]
- Fründ C., Priefert H., Steinbüchel A., Schlegel H. G. Biochemical and genetic analyses of acetoin catabolism in Alcaligenes eutrophus. J Bacteriol. 1989 Dec;171(12):6539–6548. doi: 10.1128/jb.171.12.6539-6548.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grundy F. J., Waters D. A., Takova T. Y., Henkin T. M. Identification of genes involved in utilization of acetate and acetoin in Bacillus subtilis. Mol Microbiol. 1993 Oct;10(2):259–271. doi: 10.1111/j.1365-2958.1993.tb01952.x. [DOI] [PubMed] [Google Scholar]
- Grunstein M., Hogness D. S. Colony hybridization: a method for the isolation of cloned DNAs that contain a specific gene. Proc Natl Acad Sci U S A. 1975 Oct;72(10):3961–3965. doi: 10.1073/pnas.72.10.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hanna Z., Fregeau C., Préfontaine G., Brousseau R. Construction of a family of universal expression plasmid vectors. Gene. 1984 Oct;30(1-3):247–250. doi: 10.1016/0378-1119(84)90128-8. [DOI] [PubMed] [Google Scholar]
- Hawkins C. F., Borges A., Perham R. N. A common structural motif in thiamin pyrophosphate-binding enzymes. FEBS Lett. 1989 Sep 11;255(1):77–82. doi: 10.1016/0014-5793(89)81064-6. [DOI] [PubMed] [Google Scholar]
- Hawkins C. F., Borges A., Perham R. N. Cloning and sequence analysis of the genes encoding the alpha and beta subunits of the E1 component of the pyruvate dehydrogenase multienzyme complex of Bacillus stearothermophilus. Eur J Biochem. 1990 Jul 31;191(2):337–346. doi: 10.1111/j.1432-1033.1990.tb19128.x. [DOI] [PubMed] [Google Scholar]
- Hawley D. K., McClure W. R. Compilation and analysis of Escherichia coli promoter DNA sequences. Nucleic Acids Res. 1983 Apr 25;11(8):2237–2255. doi: 10.1093/nar/11.8.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemilä H. Lipoamide dehydrogenase of Staphylococcus aureus: nucleotide sequence and sequence analysis. Biochim Biophys Acta. 1991 Dec 2;1129(1):119–123. doi: 10.1016/0167-4781(91)90225-b. [DOI] [PubMed] [Google Scholar]
- Hemilä H., Palva A., Paulin L., Arvidson S., Palva I. Secretory S complex of Bacillus subtilis: sequence analysis and identity to pyruvate dehydrogenase. J Bacteriol. 1990 Sep;172(9):5052–5063. doi: 10.1128/jb.172.9.5052-5063.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hewick R. M., Hunkapiller M. W., Hood L. E., Dreyer W. J. A gas-liquid solid phase peptide and protein sequenator. J Biol Chem. 1981 Aug 10;256(15):7990–7997. [PubMed] [Google Scholar]
- Hu C. W., Lau K. S., Griffin T. A., Chuang J. L., Fisher C. W., Cox R. P., Chuang D. T. Isolation and sequencing of a cDNA encoding the decarboxylase (E1)alpha precursor of bovine branched-chain alpha-keto acid dehydrogenase complex. Expression of E1 alpha mRNA and subunit in maple-syrup-urine-disease and 3T3-L1 cells. J Biol Chem. 1988 Jun 25;263(18):9007–9014. [PubMed] [Google Scholar]
- Huh T. L., Casazza J. P., Huh J. W., Chi Y. T., Song B. J. Characterization of two cDNA clones for pyruvate dehydrogenase E1 beta subunit and its regulation in tricarboxylic acid cycle-deficient fibroblast. J Biol Chem. 1990 Aug 5;265(22):13320–13326. [PubMed] [Google Scholar]
- Joerger R. D., Jacobson M. R., Bishop P. E. Two nifA-like genes required for expression of alternative nitrogenases by Azotobacter vinelandii. J Bacteriol. 1989 Jun;171(6):3258–3267. doi: 10.1128/jb.171.6.3258-3267.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K. R., Komuniecki R., Sun Y., Wheelock M. J. Characterization of cDNA clones for the alpha subunit of pyruvate dehydrogenase from Ascaris suum. Mol Biochem Parasitol. 1992 Mar;51(1):37–47. doi: 10.1016/0166-6851(92)90198-s. [DOI] [PubMed] [Google Scholar]
- Krüger N., Steinbüchel A. Identification of acoR, a regulatory gene for the expression of genes essential for acetoin catabolism in Alcaligenes eutrophus H16. J Bacteriol. 1992 Jul;174(13):4391–4400. doi: 10.1128/jb.174.13.4391-4400.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- López J. M., Thoms B., Rehbein H. Acetoin degradation in Bacillus subtilis by direct oxidative cleavage. Eur J Biochem. 1975 Sep 15;57(2):425–430. doi: 10.1111/j.1432-1033.1975.tb02317.x. [DOI] [PubMed] [Google Scholar]
- Matuda S., Nakano K., Ohta S., Saheki T., Kawanishi Y., Miyata T. The alpha-ketoacid dehydrogenase complexes. Sequence similarity of rat pyruvate dehydrogenase with Escherichia coli and Azotobacter vinelandii alpha-ketoglutarate dehydrogenase. Biochim Biophys Acta. 1991 May 2;1089(1):1–7. doi: 10.1016/0167-4781(91)90076-x. [DOI] [PubMed] [Google Scholar]
- McCarthy J. E., Gualerzi C. Translational control of prokaryotic gene expression. Trends Genet. 1990 Mar;6(3):78–85. doi: 10.1016/0168-9525(90)90098-q. [DOI] [PubMed] [Google Scholar]
- Niu X. D., Browning K. S., Behal R. H., Reed L. J. Cloning and nucleotide sequence of the gene for dihydrolipoamide acetyltransferase from Saccharomyces cerevisiae. Proc Natl Acad Sci U S A. 1988 Oct;85(20):7546–7550. doi: 10.1073/pnas.85.20.7546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni Y., Mitsubuchi H., Endo F., Akaboshi I., Asaka J., Matsuda I. Maple syrup urine disease. Complete primary structure of the E1 beta subunit of human branched chain alpha-ketoacid dehydrogenase complex deduced from the nucleotide sequence and a gene analysis of patients with this disease. J Clin Invest. 1990 Jul;86(1):242–247. doi: 10.1172/JCI114690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nobukuni Y., Mitsubuchi H., Endo F., Asaka J., Oyama R., Titani K., Matsuda I. Isolation and characterization of a complementary DNA clone coding for the E1 beta subunit of the bovine branched-chain alpha-ketoacid dehydrogenase complex: complete amino acid sequence of the precursor protein and its proteolytic processing. Biochemistry. 1990 Feb 6;29(5):1154–1160. doi: 10.1021/bi00457a009. [DOI] [PubMed] [Google Scholar]
- Oppermann F. B., Schmidt B., Steinbüchel A. Purification and characterization of acetoin:2,6-dichlorophenolindophenol oxidoreductase, dihydrolipoamide dehydrogenase, and dihydrolipoamide acetyltransferase of the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J Bacteriol. 1991 Jan;173(2):757–767. doi: 10.1128/jb.173.2.757-767.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oppermann F. B., Steinbüchel A. Identification and molecular characterization of the aco genes encoding the Pelobacter carbinolicus acetoin dehydrogenase enzyme system. J Bacteriol. 1994 Jan;176(2):469–485. doi: 10.1128/jb.176.2.469-485.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osterburg G., Glatting K. H., Buchert J., Wolters J. A fast method for arranging DNA sequence fragments. Comput Programs Biomed. 1983 Feb-Apr;16(1-2):61–69. doi: 10.1016/0010-468x(83)90010-7. [DOI] [PubMed] [Google Scholar]
- Palmer J. A., Madhusudhan K. T., Hatter K., Sokatch J. R. Cloning, sequence and transcriptional analysis of the structural gene for LPD-3, the third lipoamide dehydrogenase of Pseudomonas putida. Eur J Biochem. 1991 Dec 5;202(2):231–240. doi: 10.1111/j.1432-1033.1991.tb16367.x. [DOI] [PubMed] [Google Scholar]
- Perham R. N., Packman L. C. 2-Oxo acid dehydrogenase multienzyme complexes: domains, dynamics, and design. Ann N Y Acad Sci. 1989;573:1–20. doi: 10.1111/j.1749-6632.1989.tb14983.x. [DOI] [PubMed] [Google Scholar]
- Perham R. N., Packman L. C., Radford S. E. 2-Oxo acid dehydrogenase multi-enzyme complexes: in the beginning and halfway there. Biochem Soc Symp. 1987;54:67–81. [PubMed] [Google Scholar]
- Priefert H., Hein S., Krüger N., Zeh K., Schmidt B., Steinbüchel A. Identification and molecular characterization of the Alcaligenes eutrophus H16 aco operon genes involved in acetoin catabolism. J Bacteriol. 1991 Jul;173(13):4056–4071. doi: 10.1128/jb.173.13.4056-4071.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priefert H., Krüger N., Jendrossek D., Schmidt B., Steinbüchel A. Identification and molecular characterization of the gene coding for acetaldehyde dehydrogenase II (acoD) of Alcaligenes eutrophus. J Bacteriol. 1992 Feb;174(3):899–907. doi: 10.1128/jb.174.3.899-907.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priefert H., Steinbüchel A. Identification and molecular characterization of the acetyl coenzyme A synthetase gene (acoE) of Alcaligenes eutrophus. J Bacteriol. 1992 Oct;174(20):6590–6599. doi: 10.1128/jb.174.20.6590-6599.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed L. J., Hackert M. L. Structure-function relationships in dihydrolipoamide acyltransferases. J Biol Chem. 1990 Jun 5;265(16):8971–8974. [PubMed] [Google Scholar]
- Reitzer L. J., Magasanik B. Expression of glnA in Escherichia coli is regulated at tandem promoters. Proc Natl Acad Sci U S A. 1985 Apr;82(7):1979–1983. doi: 10.1073/pnas.82.7.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodriguez H., Kohr W. J., Harkins R. N. Design and operation of a completely automated Beckman microsequencer. Anal Biochem. 1984 Aug 1;140(2):538–547. doi: 10.1016/0003-2697(84)90205-7. [DOI] [PubMed] [Google Scholar]
- SAITO H., MIURA K. I. PREPARATION OF TRANSFORMING DEOXYRIBONUCLEIC ACID BY PHENOL TREATMENT. Biochim Biophys Acta. 1963 Aug 20;72:619–629. [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stegemann H., Francksen H., Macko V. Potato proteins: genetic and physiological changes, evaluated by one- and two-dimensional PAA-gel-techniques. Z Naturforsch C. 1973 Nov-Dec;28(11):722–732. doi: 10.1515/znc-1973-11-1213. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the dihydrolipoamide acetyltransferase component. Eur J Biochem. 1983 Jul 1;133(3):481–489. doi: 10.1111/j.1432-1033.1983.tb07490.x. [DOI] [PubMed] [Google Scholar]
- Stephens P. E., Darlison M. G., Lewis H. M., Guest J. R. The pyruvate dehydrogenase complex of Escherichia coli K12. Nucleotide sequence encoding the pyruvate dehydrogenase component. Eur J Biochem. 1983 Jun 1;133(1):155–162. doi: 10.1111/j.1432-1033.1983.tb07441.x. [DOI] [PubMed] [Google Scholar]
- Strauss E. C., Kobori J. A., Siu G., Hood L. E. Specific-primer-directed DNA sequencing. Anal Biochem. 1986 Apr;154(1):353–360. doi: 10.1016/0003-2697(86)90536-1. [DOI] [PubMed] [Google Scholar]
- Szeto W. W., Nixon B. T., Ronson C. W., Ausubel F. M. Identification and characterization of the Rhizobium meliloti ntrC gene: R. meliloti has separate regulatory pathways for activation of nitrogen fixation genes in free-living and symbiotic cells. J Bacteriol. 1987 Apr;169(4):1423–1432. doi: 10.1128/jb.169.4.1423-1432.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thöny B., Hennecke H. The -24/-12 promoter comes of age. FEMS Microbiol Rev. 1989 Dec;5(4):341–357. doi: 10.1016/0168-6445(89)90028-4. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Towbin H., Staehelin T., Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B., Gillespie D. Preparative and analytical purification of DNA from agarose. Proc Natl Acad Sci U S A. 1979 Feb;76(2):615–619. doi: 10.1073/pnas.76.2.615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wallbrandt P., Tegman V., Jonsson B. H., Wieslander A. Identification and analysis of the genes coding for the putative pyruvate dehydrogenase enzyme complex in Acholeplasma laidlawii. J Bacteriol. 1992 Feb;174(4):1388–1396. doi: 10.1128/jb.174.4.1388-1396.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang G. F., Kuriki T., Roy K. L., Kaneda T. The primary structure of branched-chain alpha-oxo acid dehydrogenase from Bacillus subtilis and its similarity to other alpha-oxo acid dehydrogenases. Eur J Biochem. 1993 May 1;213(3):1091–1099. doi: 10.1111/j.1432-1033.1993.tb17858.x. [DOI] [PubMed] [Google Scholar]
- Wexler I. D., Hemalatha S. G., Patel M. S. Sequence conservation in the alpha and beta subunits of pyruvate dehydrogenase and its similarity to branched-chain alpha-keto acid dehydrogenase. FEBS Lett. 1991 Apr 22;282(1):209–213. doi: 10.1016/0014-5793(91)80479-m. [DOI] [PubMed] [Google Scholar]
- Wheelock M. J., Komuniecki R., Duran E., Johnson K. R. Characterization of cDNA clones for the beta subunit of pyruvate dehydrogenase from Ascaris suum. Mol Biochem Parasitol. 1991 Mar;45(1):9–17. doi: 10.1016/0166-6851(91)90022-x. [DOI] [PubMed] [Google Scholar]
- Widdel F., Pfennig N. Studies on dissimilatory sulfate-reducing bacteria that decompose fatty acids. I. Isolation of new sulfate-reducing bacteria enriched with acetate from saline environments. Description of Desulfobacter postgatei gen. nov., sp. nov. Arch Microbiol. 1981 Jul;129(5):395–400. doi: 10.1007/BF00406470. [DOI] [PubMed] [Google Scholar]
- Yang C. C., Nash H. A. The interaction of E. coli IHF protein with its specific binding sites. Cell. 1989 Jun 2;57(5):869–880. doi: 10.1016/0092-8674(89)90801-5. [DOI] [PubMed] [Google Scholar]
- de Lorenzo V., Herrero M., Metzke M., Timmis K. N. An upstream XylR- and IHF-induced nucleoprotein complex regulates the sigma 54-dependent Pu promoter of TOL plasmid. EMBO J. 1991 May;10(5):1159–1167. doi: 10.1002/j.1460-2075.1991.tb08056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]