Abstract
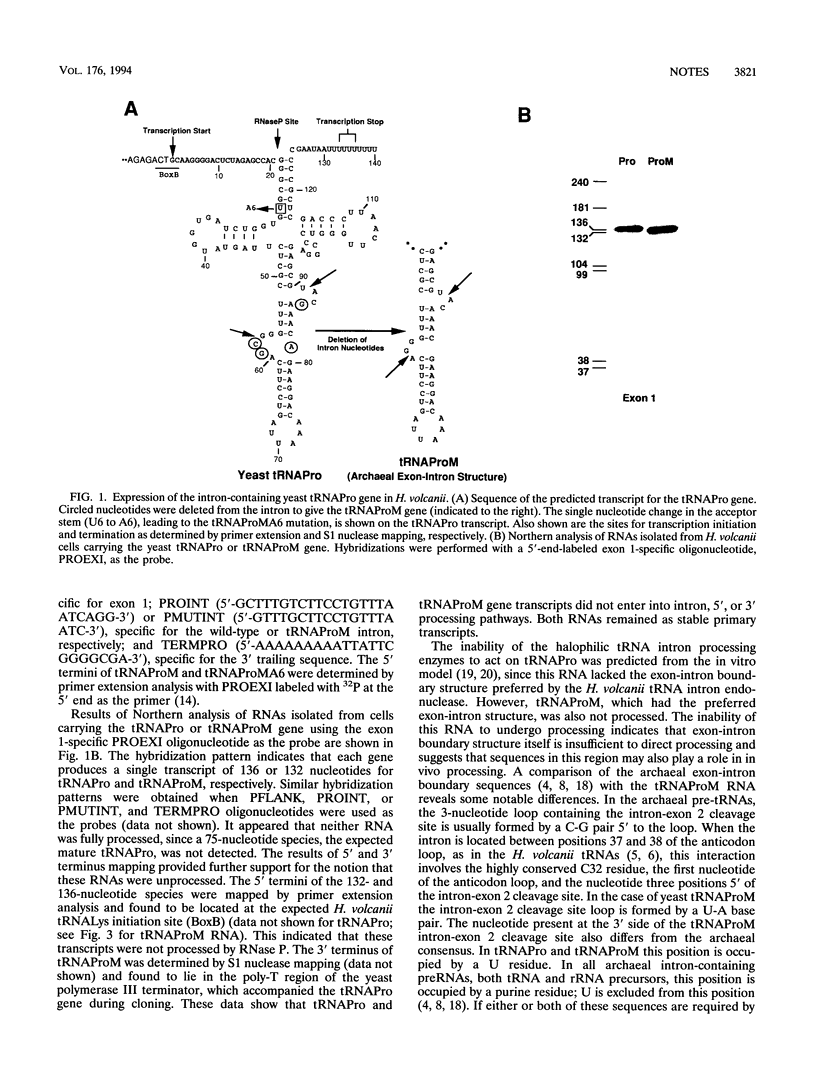
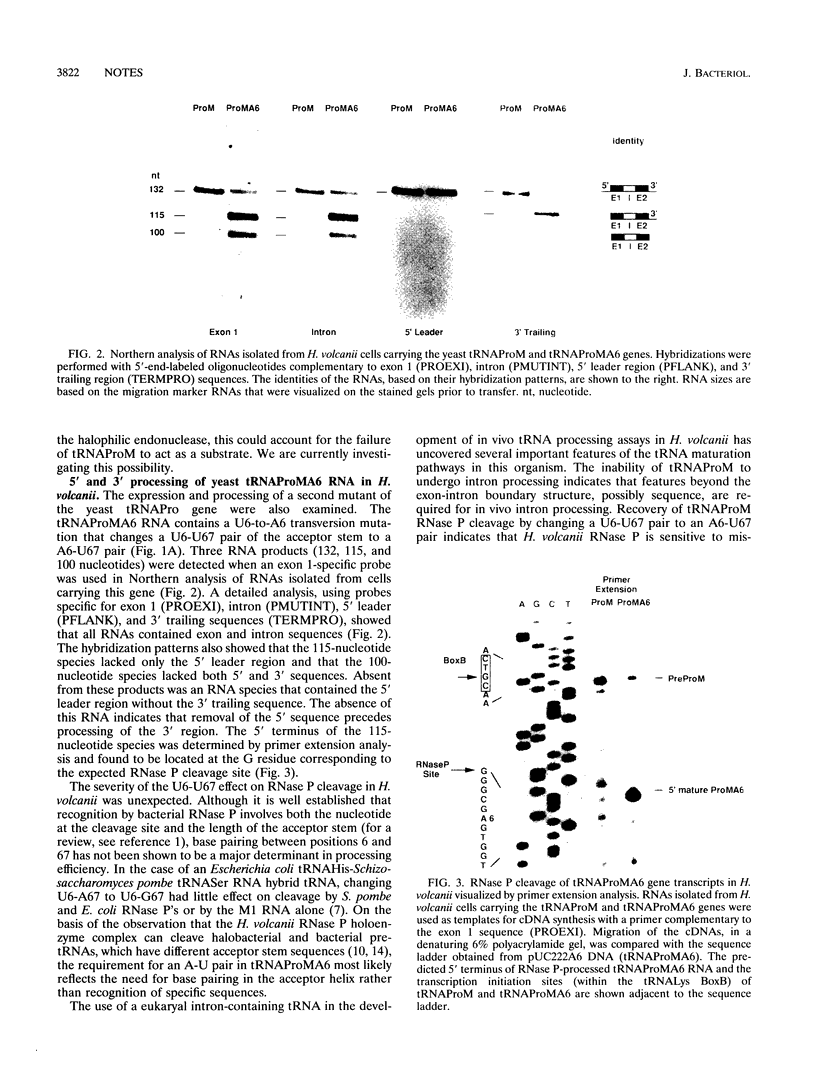
Expression of the yeast tRNAPro(UGG) gene in Haloferax volcanii resulted in the production of a single stable transcript that had not undergone intron processing or processing of 5' and 3' flanking sequences. Mutation of the exon-intron boundary region of this RNA to produce a precursor RNA with the preferred halobacterial consensus exon-intron boundary structure did not restore intron processing. Processing of 5' and 3' flanking sequences was restored when the acceptor stem U6-U67 pair was changed to A6-U67. The significance of these results in defining the recognition requirements of tRNA maturation enzymes in the halophilic domain Archaea is discussed.
Full text
PDF

Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Altman S., Kirsebom L., Talbot S. Recent studies of ribonuclease P. FASEB J. 1993 Jan;7(1):7–14. doi: 10.1096/fasebj.7.1.7916700. [DOI] [PubMed] [Google Scholar]
- Baldi M. I., Mattoccia E., Bufardeci E., Fabbri S., Tocchini-Valentini G. P. Participation of the intron in the reaction catalyzed by the Xenopus tRNA splicing endonuclease. Science. 1992 Mar 13;255(5050):1404–1408. doi: 10.1126/science.1542788. [DOI] [PubMed] [Google Scholar]
- Bufardeci E., Fabbri S., Baldi M. I., Mattoccia E., Tocchini-Valentini G. P. In vitro genetic analysis of the structural features of the pre-tRNA required for determination of the 3' splice site in the intron excision reaction. EMBO J. 1993 Dec;12(12):4697–4704. doi: 10.1002/j.1460-2075.1993.tb06158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalgaard J. Z., Garrett R. A. Protein-coding introns from the 23S rRNA-encoding gene form stable circles in the hyperthermophilic archaeon Pyrobaculum organotrophum. Gene. 1992 Nov 2;121(1):103–110. doi: 10.1016/0378-1119(92)90167-n. [DOI] [PubMed] [Google Scholar]
- Daniels C. J., Gupta R., Doolittle W. F. Transcription and excision of a large intron in the tRNATrp gene of an archaebacterium, Halobacterium volcanii. J Biol Chem. 1985 Mar 10;260(5):3132–3134. [PubMed] [Google Scholar]
- Datta P. K., Hawkins L. K., Gupta R. Presence of an intron in elongator methionine-tRNA of Halobacterium volcanii. Can J Microbiol. 1989 Jan;35(1):189–194. doi: 10.1139/m89-029. [DOI] [PubMed] [Google Scholar]
- Holm P. S., Krupp G. The acceptor stem in pre-tRNAs determines the cleavage specificity of RNase P. Nucleic Acids Res. 1992 Feb 11;20(3):421–423. doi: 10.1093/nar/20.3.421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjems J., Jensen J., Olesen T., Garrett R. A. Comparison of transfer RNA and ribosomal RNA intron splicing in the extreme thermophile and archaebacterium Desulfurococcus mobilis. Can J Microbiol. 1989 Jan;35(1):210–214. doi: 10.1139/m89-033. [DOI] [PubMed] [Google Scholar]
- Lawrence N., Wesolowski D., Gold H., Bartkiewicz M., Guerrier-Takada C., McClain W. H., Altman S. Characteristics of ribonuclease P from various organisms. Cold Spring Harb Symp Quant Biol. 1987;52:233–238. doi: 10.1101/sqb.1987.052.01.028. [DOI] [PubMed] [Google Scholar]
- Mattoccia E., Baldi I. M., Gandini-Attardi D., Ciafrè S., Tocchini-Valentini G. P. Site selection by the tRNA splicing endonuclease of Xenopus laevis. Cell. 1988 Nov 18;55(4):731–738. doi: 10.1016/0092-8674(88)90231-0. [DOI] [PubMed] [Google Scholar]
- Nieuwlandt D. T., Carr M. B., Daniels C. J. In vivo processing of an intron-containing archael tRNA. Mol Microbiol. 1993 Apr;8(1):93–99. doi: 10.1111/j.1365-2958.1993.tb01206.x. [DOI] [PubMed] [Google Scholar]
- Nieuwlandt D. T., Daniels C. J. An expression vector for the archaebacterium Haloferax volcanii. J Bacteriol. 1990 Dec;172(12):7104–7110. doi: 10.1128/jb.172.12.7104-7110.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nieuwlandt D. T., Haas E. S., Daniels C. J. The RNA component of RNase P from the archaebacterium Haloferax volcanii. J Biol Chem. 1991 Mar 25;266(9):5689–5695. [PubMed] [Google Scholar]
- Ogden R. C., Lee M. C., Knapp G. Transfer RNA splicing in Saccharomyces cerevisiae: defining the substrates. Nucleic Acids Res. 1984 Dec 21;12(24):9367–9382. doi: 10.1093/nar/12.24.9367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reyes V. M., Abelson J. Substrate recognition and splice site determination in yeast tRNA splicing. Cell. 1988 Nov 18;55(4):719–730. doi: 10.1016/0092-8674(88)90230-9. [DOI] [PubMed] [Google Scholar]
- Shapero M. H., Greer C. L. Exon sequence and structure requirements for tRNA splicing in Saccharomyces cerevisiae. Biochemistry. 1992 Mar 3;31(8):2359–2367. doi: 10.1021/bi00123a022. [DOI] [PubMed] [Google Scholar]
- Thompson L. D., Brandon L. D., Nieuwlandt D. T., Daniels C. J. Transfer RNA intron processing in the halophilic archaebacteria. Can J Microbiol. 1989 Jan;35(1):36–42. doi: 10.1139/m89-006. [DOI] [PubMed] [Google Scholar]
- Thompson L. D., Daniels C. J. A tRNA(Trp) intron endonuclease from Halobacterium volcanii. Unique substrate recognition properties. J Biol Chem. 1988 Dec 5;263(34):17951–17959. [PubMed] [Google Scholar]
- Thompson L. D., Daniels C. J. Recognition of exon-intron boundaries by the Halobacterium volcanii tRNA intron endonuclease. J Biol Chem. 1990 Oct 25;265(30):18104–18111. [PubMed] [Google Scholar]
- Turmel M., Boulanger J., Schnare M. N., Gray M. W., Lemieux C. Six group I introns and three internal transcribed spacers in the chloroplast large subunit ribosomal RNA gene of the green alga Chlamydomonas eugametos. J Mol Biol. 1991 Mar 20;218(2):293–311. doi: 10.1016/0022-2836(91)90713-g. [DOI] [PubMed] [Google Scholar]