Abstract
The transcriptional organization of the exoenzyme S trans-regulatory locus was studied by using promoter fusion and transcriptional start site mapping analyses. The 5' regions flanking open reading frames encoding ExsC, ExsB, ExsA, and ExsD were cloned in both orientations into the promoter vector pQF26, which contains the chloramphenicol acetyltransferase reporter gene (cat). CAT activity from each promoter fusion transformed into Pseudomonas aeruginosa and Escherichia coli was measured. The trans-regulatory locus promoters demonstrated low to undetectable CAT activity in E. coli regardless of the orientation of the DNA fragment relative to the reporter gene. In P. aeruginosa two of the promoter clones containing DNA located 5' of exsC (pC) and exsD (pD) demonstrated significant CAT activity. Transcriptional initiation from pC and pD was dependent on the orientation of the DNA fragment, the inclusion of a chelator in the growth medium, and the presence of a functional exsA gene. Transcriptional start sites were mapped for the pC and pD promoter regions by using total RNA isolated from P. aeruginosa strains grown in medium including a chelator. Our data are consistent with an operon model for the transcriptional organization of the exoenzyme S trans-regulatory locus. In addition, ExsA appears to be involved in controlling transcriptional initiation from both the trans-regulatory locus and a region located immediately downstream of the exsA gene.
Full text
PDF


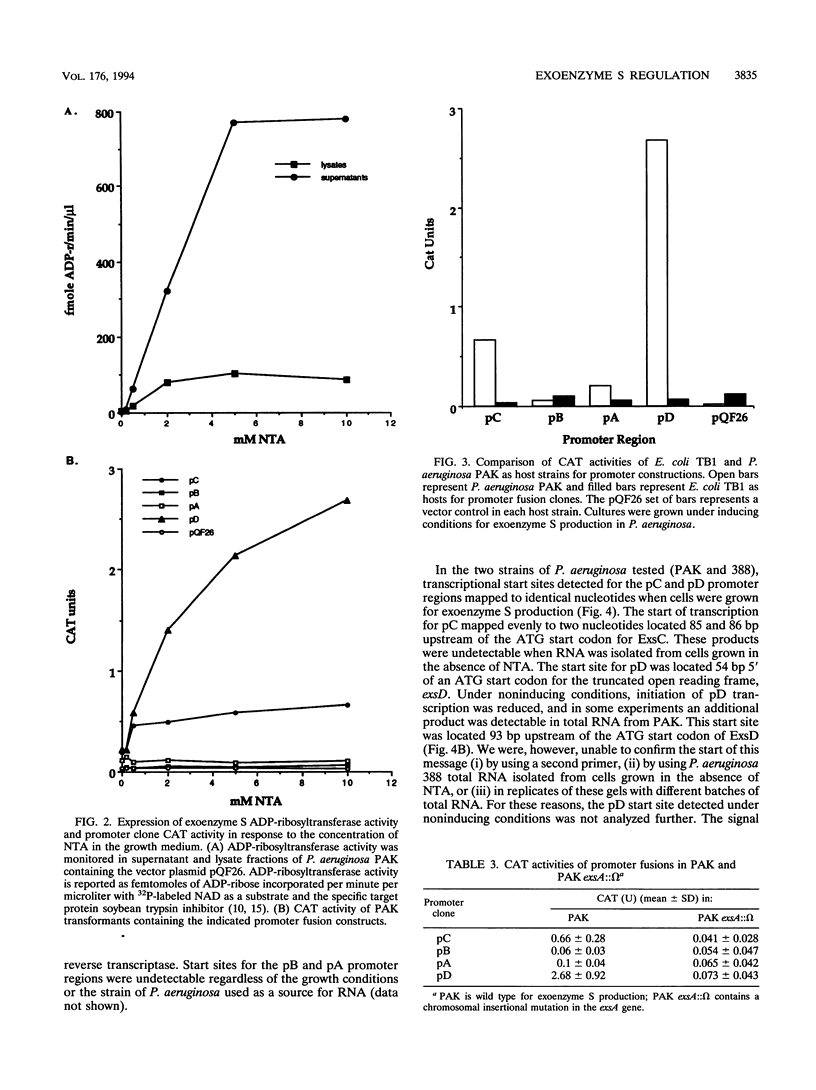
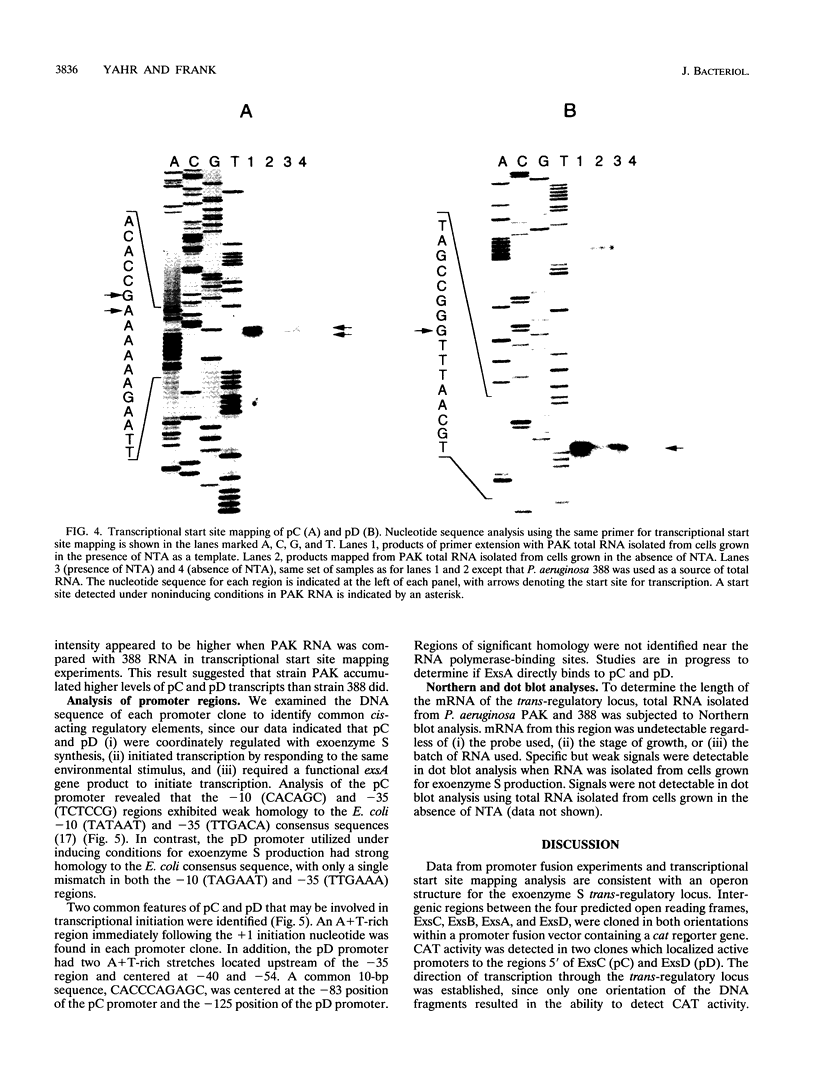


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bjorn M. J., Pavlovskis O. R., Thompson M. R., Iglewski B. H. Production of exoenzyme S during Pseudomonas aeruginosa infections of burned mice. Infect Immun. 1979 Jun;24(3):837–842. doi: 10.1128/iai.24.3.837-842.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer H. W., Roulland-Dussoix D. A complementation analysis of the restriction and modification of DNA in Escherichia coli. J Mol Biol. 1969 May 14;41(3):459–472. doi: 10.1016/0022-2836(69)90288-5. [DOI] [PubMed] [Google Scholar]
- Coburn J., Dillon S. T., Iglewski B. H., Gill D. M. Exoenzyme S of Pseudomonas aeruginosa ADP-ribosylates the intermediate filament protein vimentin. Infect Immun. 1989 Mar;57(3):996–998. doi: 10.1128/iai.57.3.996-998.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coburn J., Kane A. V., Feig L., Gill D. M. Pseudomonas aeruginosa exoenzyme S requires a eukaryotic protein for ADP-ribosyltransferase activity. J Biol Chem. 1991 Apr 5;266(10):6438–6446. [PubMed] [Google Scholar]
- Coburn J. Pseudomonas aeruginosa exoenzyme S. Curr Top Microbiol Immunol. 1992;175:133–143. doi: 10.1007/978-3-642-76966-5_7. [DOI] [PubMed] [Google Scholar]
- Farinha M. A., Kropinski A. M. Construction of broad-host-range vectors for general cloning and promoter selection in Pseudomonas and Escherichia coli. Gene. 1989 Apr 30;77(2):205–210. doi: 10.1016/0378-1119(89)90068-1. [DOI] [PubMed] [Google Scholar]
- Frank D. W., Iglewski B. H. Cloning and sequence analysis of a trans-regulatory locus required for exoenzyme S synthesis in Pseudomonas aeruginosa. J Bacteriol. 1991 Oct;173(20):6460–6468. doi: 10.1128/jb.173.20.6460-6468.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. W., Iglewski B. H. Kinetics of toxA and regA mRNA accumulation in Pseudomonas aeruginosa. J Bacteriol. 1988 Oct;170(10):4477–4483. doi: 10.1128/jb.170.10.4477-4483.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank D. W., Nair G., Schweizer H. P. Construction and characterization of chromosomal insertional mutations of the Pseudomonas aeruginosa exoenzyme S trans-regulatory locus. Infect Immun. 1994 Feb;62(2):554–563. doi: 10.1128/iai.62.2.554-563.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H., Coburn J., Collier R. J. The eukaryotic host factor that activates exoenzyme S of Pseudomonas aeruginosa is a member of the 14-3-3 protein family. Proc Natl Acad Sci U S A. 1993 Mar 15;90(6):2320–2324. doi: 10.1073/pnas.90.6.2320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Hoe N. P., Goguen J. D. Temperature sensing in Yersinia pestis: translation of the LcrF activator protein is thermally regulated. J Bacteriol. 1993 Dec;175(24):7901–7909. doi: 10.1128/jb.175.24.7901-7909.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iglewski B. H., Sadoff J., Bjorn M. J., Maxwell E. S. Pseudomonas aeruginosa exoenzyme S: an adenosine diphosphate ribosyltransferase distinct from toxin A. Proc Natl Acad Sci U S A. 1978 Jul;75(7):3211–3215. doi: 10.1073/pnas.75.7.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulich S. M., Frank D. W., Barbieri J. T. Purification and characterization of exoenzyme S from Pseudomonas aeruginosa 388. Infect Immun. 1993 Jan;61(1):307–313. doi: 10.1128/iai.61.1.307-313.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure W. R. Mechanism and control of transcription initiation in prokaryotes. Annu Rev Biochem. 1985;54:171–204. doi: 10.1146/annurev.bi.54.070185.001131. [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Bradley J., Lochner J. E., Iglewski B. H. The role of exoenzyme S in infections with Pseudomonas aeruginosa. J Infect Dis. 1985 Oct;152(4):716–721. doi: 10.1093/infdis/152.4.716. [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Iglewski B. H. Contribution of exoenzyme S to the virulence of Pseudomonas aeruginosa. Antibiot Chemother (1971) 1985;36:40–48. doi: 10.1159/000410470. [DOI] [PubMed] [Google Scholar]
- Nicas T. I., Iglewski B. H. Isolation and characterization of transposon-induced mutants of Pseudomonas aeruginosa deficient in production of exoenzyme S. Infect Immun. 1984 Aug;45(2):470–474. doi: 10.1128/iai.45.2.470-474.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen R. H., DeBusscher G., McCombie W. R. Development of broad-host-range vectors and gene banks: self-cloning of the Pseudomonas aeruginosa PAO chromosome. J Bacteriol. 1982 Apr;150(1):60–69. doi: 10.1128/jb.150.1.60-69.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross W., Gosink K. K., Salomon J., Igarashi K., Zou C., Ishihama A., Severinov K., Gourse R. L. A third recognition element in bacterial promoters: DNA binding by the alpha subunit of RNA polymerase. Science. 1993 Nov 26;262(5138):1407–1413. doi: 10.1126/science.8248780. [DOI] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey D. G., Frank D. W., Farinha M. A., Kropinski A. M., Iglewski B. H. Multiple promoters control the regulation of the Pseudomonas aeruginosa regA gene. Mol Microbiol. 1990 Mar;4(3):499–503. doi: 10.1111/j.1365-2958.1990.tb00616.x. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- von Gabain A., Belasco J. G., Schottel J. L., Chang A. C., Cohen S. N. Decay of mRNA in Escherichia coli: investigation of the fate of specific segments of transcripts. Proc Natl Acad Sci U S A. 1983 Feb;80(3):653–657. doi: 10.1073/pnas.80.3.653. [DOI] [PMC free article] [PubMed] [Google Scholar]