Abstract
Certain chemokine receptors serve as cofactors for HIV type 1 envelope (env)-mediated cell–cell fusion and virus infection of CD4-positive cells. Macrophage tropic (M-tropic) HIV-1 isolates use CCR5, and T cell tropic (T-tropic) strains use CXCR4. To investigate the cofactors used by simian immunodeficiency viruses (SIV), we tested four T-tropic and two M-tropic SIV env proteins for their ability to mediate cell–cell fusion with cells expressing CD4 and either human or nonhuman primate chemokine receptors. Unlike HIV-1, both M- and T-tropic SIV envs used CCR5 but not CXCR4 or the other chemokine receptors tested. However, by testing a panel of CCR5/CCR2b chimeras, we found that the structural requirements for CCR5 utilization by M-tropic and T-tropic SIV strains were different. T-tropic SIV strains required the second extracellular loop of CCR5 whereas a closely related M-tropic SIV strain could, like M-tropic HIV-1 strains, use the amino-terminal domain of CCR5. As few as two amino acid changes in the SIV env V3 domain affected the regions of CCR5 that were critical for fusogenic activity. Receptor signaling was not required for either fusion or infection. Our results suggest that viral tropism may be influenced not only by the coreceptors used by a given virus strain but also by how a given coreceptor is used.
For HIV type 1 (HIV-1) to infect a cell, the viral envelope (env) protein must bind to CD4 and mediate fusion between the viral envelope and host cell membrane. Fusion occurs only when both CD4 and an appropriate coreceptor are expressed on the host cell surface; CD4 alone is not sufficient for viral infection. Recently, members of the chemokine receptor family have been shown to serve as HIV-1 coreceptors (1–7). Macrophage tropic (M-tropic) strains of HIV-1 use the chemokine receptor CCR5, and T cell tropic (T-tropic) strains require the expression of CXCR4 in conjunction with CD4 for membrane fusion and infection to occur. In addition, other chemokine receptors such as CCR2b and CCR3 can function as coreceptors for some viruses, and some viruses can use more than one chemokine receptor (3, 5, 8).
The chemokine receptor repertoire used by a given virus strain plays an important role in governing viral tropism. M-tropic HIV-1 strains, which use CCR5, are involved in sexual transmission and are the predominant virus type found during the asymptomatic period (9–15). The critical role of CCR5 in HIV infection and transmission is demonstrated by the finding that a 32-bp deletion in CCR5 that effectively renders ≈1% of Caucasians CCR5-negative also confers a high degree of resistance to virus infection (16–19). With time, through the accumulation of amino acid changes in the viral env protein, T-tropic viruses may emerge that can use CXCR4 as a cofactor. Acquisition of the ability to use additional chemokine receptors may broaden viral host range and enable the virus to evade selective pressures against CCR5 usage, such as high levels of the CCR5 ligands RANTES, MIP-1α, and MIP-1β, which have anti-viral properties (20, 21).
Recent work has shown that T-tropic HIV-1 env proteins can directly interact with CXCR4 (22) whereas M-tropic env proteins interact with CCR5 (23, 24). Interactions with CCR5 are conformationally complex and involve multiple CCR5 domains (25, 26). To study the role of chemokine receptors in simian immunodeficiency virus (SIV) tropism and pathogenesis, we examined the ability of SIV env proteins derived from viruses with distinct cell tropisms to use human and nonhuman primate chemokine receptors in cell–cell fusion assays. Like HIV-1, M-tropic SIV strains (which replicate in rhesus macrophages) used CCR5. However, unlike HIV-1, the T-tropic SIV strains studied here used CCR5 but not human or rhesus CXCR4. Of interest, differences were observed in the use of CCR5 domains, with an M-tropic SIV env protein interacting with CCR5 in a manner similar to that seen with M-tropic HIV-1 env proteins; T-tropic SIV env proteins were dependent on the second extracellular loop of CCR5. Sequence changes in the V3 loop [which, in HIV-1, plays a major role in governing cell tropism and chemokine receptor usage (3, 27–29)] were found to affect the ability of SIV env to use the amino-terminal domain of CCR5. Finally, receptor signaling was not required for either virus infection or SIV env-mediated cell–cell fusion.
MATERIALS AND METHODS
Plasmid Constructs.
Human CD4 and all human chemokine receptors used were cloned into pCDNA3 under the control of the CMV and T7 promoters. Cloning of the nonhuman primate CCR5 and CXCR4 homologs and CCR5 mutants will be described elsewhere (unpublished work). The CCR5/CCR2b chimeric receptors, also cloned into pCDNA3, have been described (25). The SIV env clones B670-Cl3 (30) and SIVmac17E-Fr (32) were cloned into pCDNA3 for use in the cell–cell fusion assay.
Cells and Viruses.
HeLa cells were maintained in DMEM supplemented with 10% calf serum (DMEM-10). The Japanese quail fibrosarcoma cell line QT6-C5 was maintained in DMEM-10 or M199 supplemented with 10% tryptose phosphate broth, 5% fetal calf serum, and 1% heat inactivated chicken serum. Recombinant vaccinia viruses were used to express the SIV and HIV-1 env indicated in parentheses: v194 (SIVmac251), vCB74 (SIVmac239), vCB75 (SIVmac316), vCB76 (SIVmac316mut) (C.C.B. and E. A. Berger, unpublished work), vCB28 (HIV-1 JR-FL) (33), and vSC60 (HIV-1 IIIB, BH8 clone) (S. Chakrabarti and B. Moss, personal communication). The recombinant vaccinia virus vTF1.1 was used to express T7 RNA polymerase (34).
Gene Reporter Fusion Assay.
We used a modified version of the gene reporter fusion assay described by Nussbaum et al. to quantitate cell–cell fusion (5, 25, 35). In summary, SIV or HIV-1 env proteins and T7 RNA polymerase were expressed in HeLa cells by infection with recombinant vaccinia viruses, incubated overnight at 32°C, and mixed with quail QT6 cells transfected the previous day with plasmids encoding CD4, a chemokine receptor, and luciferase under control of the T7 promoter. After 8–10 h of incubation at 37°C, cells were lysed, and luciferase activity was measured. For fusion assays using the SIV env B670-Cl3 and SIVmac17E-Fr, HeLa effector cells were infected for 30–45 minutes with vTF1.1 and then transfected with the env constructs. After overnight incubation at 32°C, cells were lifted using 0.5 mM EDTA in PBS, washed, and added to target cells. Cell–cell fusion was initiated and quantitated as described above.
Infection with Luciferase Virus.
Luciferase virus stocks were prepared in 293T cells as described (36, 37) using pCDNA3-BK28 and pNL-Luc-E−R− vectors. Entry of luciferase virus was measured by transfecting U87-MG cells with the indicated cofactor and pT4 followed by overnight incubation. Cells were infected with 50 ng of BK28-luc virus (molecular clone of SIV 251) for 16–24 h before the medium was removed and replaced with 500 μl of fresh media. Cells were harvested 4 days postinfection by aspirating media and lysing cells in 150 μl of 0.5% Nonidet P-40/PBS. Luciferase activity was quantitated by measuring 20 μl of the resulting lysate.
RESULTS
Human Chemokine Receptor Utilization by SIV.
To determine whether chemokine receptors play a role in SIV fusion with target cells, we screened six SIV env proteins for the ability to mediate cell–cell fusion with cells expressing human CD4 and different human chemokine receptors using a gene reporter fusion assay (5, 35). In this assay, env protein and T7 polymerase were expressed in HeLa effector cells, and CD4, a chemokine receptor, and luciferase (under the transcriptional control of the T7 promoter) were expressed in quail QT6 cells. QT6 cells were used because they fail to support HIV-1 env-mediated cell–cell fusion when they express CD4 alone and can be transfected with high efficiency (5, 25). HeLa and QT6 cells were mixed, resulting in cytoplasmic mixing and luciferase transcription and expression only if cell–cell fusion occurred. Thus, quantitative measurement of luciferase activity provided a convenient and sensitive means to measure cell–cell fusion.
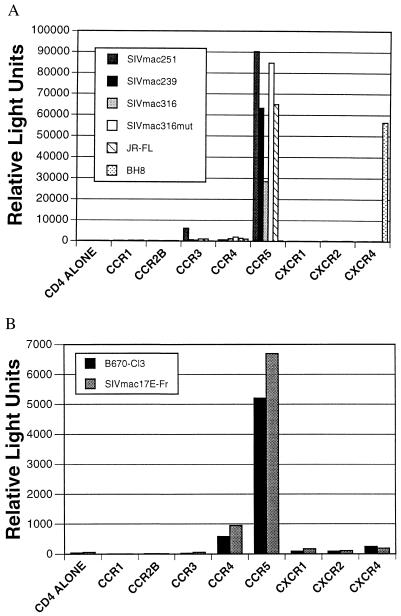
Recombinant vaccinia viruses were used to express the T-tropic env proteins derived from SIVmac251 and SIVmac239. In addition, recombinant viruses that directed the expression of a M-tropic env variant of SIVmac239 (SIVmac316) as well as a T-tropic env mutant of SIVmac316 (SIVmac316 mut) that differs from the parental env protein by only two amino acids were used (38, 39). HeLa cells were infected with a recombinant vaccinia virus encoding an env gene and with vTF1.1, which encodes T7 polymerase. After overnight incubation, the cells were mixed with QT6 cells expressing CD4 and the indicated human chemokine receptor. Cells were lysed 8 h after mixing, and luciferase activity was determined. T-tropic (BH8) and M-tropic (JR-FL) HIV-1 strains were used as positive controls. All four SIV env proteins mediated cell–cell fusion through CCR5 (Fig. 1a). In addition, SIVmac251 used CCR3 to a very limited degree. Other human chemokine receptors (CCR1, CCR2b, CCR4, CXCR1, CXCR2, and CXCR4) lacked fusion cofactor activity for all of the SIV env proteins tested. These results were surprising because CXCR4 is the major cofactor for T-tropic HIV-1 strains (1, 3, 7, 8). To extend these observations, we examined two additional, independently derived SIV env proteins: SIVmac17E-Fr, a M-tropic infectious clone constructed by replacing the viral env of SIVmac239 with a PCR-amplified env fragment derived from brain tissue from a macaque involved in a serial brain passage of SIVmac239 (32), and SIV/DeltaB670-Cl3, a genetic member of the quasispecies comprising the primary isolate SIV/DeltaB670 (30). Selective amplification of SIV/DeltaB670-Cl3 is known to occur by serial passage of the parental SIV/DeltaB670 in human T cell lines, a finding that confirms the T-tropism of this virus. Expression of these env proteins in HeLa cells by transient transfection resulted in fusion only with cells expressing CD4 and CCR5 (Fig. 1b). Thus, all four T-tropic and both M-tropic SIV env proteins mediated cell–cell fusion in a CCR5-dependent manner.
Figure 1.
Chemokine receptor use by SIV env proteins. (A) HeLa cells were infected with vTF1.1 and the indicated SIV or HIV-1 env protein. QT6 cells were transfected with plasmids encoding huCD4, the indicated chemokine receptor, and a plasmid encoding luciferase under control of the T7 promoter. After overnight incubation, HeLa and QT6 cells were mixed, incubated at 37°C for 8 h to allow fusion to occur, and lysed, and the amount of luciferase activity was determined in relative light units. (B) As in A, except that the indicated SIV env proteins were expressed transiently after transfection of HeLa cells. As a result, a much smaller fraction of HeLa cells expressed env, resulting in cell–cell fusion that was less efficient than in A (note difference in scales).
Use of Nonhuman Primate CXCR4 and CCR5 Homologs by SIV.
The fusogenic activity of simian homologs of CCR5 and CXCR4 was analyzed for the SIV env proteins of varying tropisms to determine whether the results obtained with the human receptors were a true indicator of their biologic activity. The panel of SIV env proteins was analyzed in the cell–cell fusion assay against CXCR4 and CCR5 derived from Rhesus macaque and CCR5 derived from pigtail macaque, cynomologous monkey, chimpanzee, gorilla, and baboon (Table 1). Although env derived from HIV-1 BH8 readily mediated fusion with cells expressing human CD4 and rhesus CXCR4, the SIV envs were unable to use either human or simian CXCR4 although a very weak signal was consistently obtained with SIVmac316 env. In contrast, the nonhuman primate CCR5 homologs with human CD4 all supported cell–cell fusion with the SIV env proteins tested (Table 1). Thus, in contrast to T-tropic HIV-1 strains, the T-tropic SIV strains examined here failed to use either human or rhesus CXCR4. Although human CD4 was used in these assays, the SIV strains used here replicate efficiently in CD4-positive human T cells, indicating that they can use human CD4 as a receptor for virus entry (40).
Table 1.
Use of nonhuman primate CXCR4 and CCR5 homologs by SIV
| SIVmac251 | SIVmac239 | SIVmac316 | SIVmac316mut | SIVB670-Cl3 | SIVmac17E-Fr | HIV-1 BH8 | |
|---|---|---|---|---|---|---|---|
| Human CXCR4 | − | − | − | − | − | − | +++ |
| Rhesus CXCR4 | − | − | +/− | − | − | − | +++ |
| Human CCR5 | +++ | +++ | +++ | +++ | +++ | +++ | − |
| Rhesus CCR5 | +++ | +++ | +++ | +++ | ++ | ++ | − |
| Pigtail CCR5 | +++ | +++ | +++ | +++ | +++ | +++ | − |
| Cynomologous CCR5 | ++ | ++ | +++ | +++ | + | ++ | − |
| Chimpanzee CCR5 | +++ | +++ | +++ | +++ | ++ | ++ | − |
| Gorilla CCR5 | +++ | +++ | +++ | +++ | ++ | ++ | − |
| Baboon CCR5 | +++ | ++ | +++ | +++ | +++ | +++ | − |
Symbols correspond to signal-to-noise ratios (S:N) of: −, S:N ≤ 5; +/−, 5 < S:N ≤ 10; +, 10 < S:N ≤ 20; ++, 20 < S:N ≤ 50; +++, S:N > 50.
Differential Use of Human CCR5 by T-Tropic SIV and M-Tropic HIV-1 Strains.
Despite its T-tropic phenotype, SIVmac239 enters macrophages as efficiently as M-tropic variants but fails to replicate (41). As few as five amino acid changes in the SIVmac239 env protein enable it to replicate in macrophages, indicating that changes in env can affect postentry steps of the virus life cycle (38, 41, 42). The results shown in Fig. 1 suggest that the failure of SIVmac239 to infect macrophages can not be accounted for simply by an inability to use CCR5 because both T- and M-tropic strains used this chemokine receptor. Because CCR5 is expressed in both T cells and macrophages, we sought to determine if tropism differences may result from differential use of CCR5 rather than from the use of different coreceptors. Recently, we used CCR5/CCR2b chimeras to map CCR5 domains that are important for cofactor function and found that interactions between HIV-1 and CCR5 are conformationally complex and involve multiple CCR5 domains, with the amino-terminal region and first extracellular loop being particularly important (25). In addition, we found that M-tropic HIV-1 strains used CCR5 differently than the dual-tropic strain 89.6. Therefore, to determine whether T-tropic SIV env proteins use CCR5 in a manner distinct from that seen with M-tropic HIV-1 strains, we tested their ability to mediate fusion with cells expressing CD4 and various CCR5/CCR2b chimeras (Fig. 2).
Figure 2.
CCR5/CCR2b chimeras. CCR5/CCR2b receptor chimeras were prepared as described (25). Designations used in our previous work (25) are shown in parentheses, and designations used here (5222, 2522, etc.) are shown in bold.
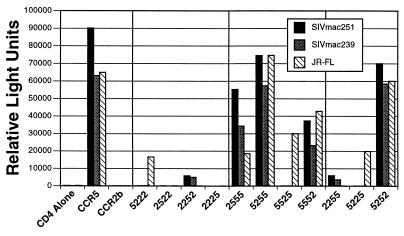
We first asked whether single CCR5 domains introduced into a CCR2b background could support cell–cell fusion by the T-tropic strains SIVmac251 and SIVmac239 and by the M-tropic HIV-1 strain JR-FL (Fig. 3). Chimera 5222, but not chimeras 2522, 2252, or 2225, supported fusion by JR-FL, indicating that the amino-terminal domain of CCR5 is the only CCR5 domain that is sufficient to confer cofactor activity to CCR2b (25). In contrast, 5222 did not support fusion by the T-tropic SIV env proteins although a low level of fusion was consistently observed when cells expressed CD4 and chimera 2252. Thus, the second extracellular loop rather than the amino-terminal domain was the only CCR5 domain that was sufficient to confer cofactor function to CCR2b for the T-tropic SIV envs. The reciprocal chimeras, in which individual CCR5 domains were replaced with corresponding CCR2b regions, showed that the second extracellular loop of CCR5 was both necessary and sufficient for T-tropic, SIV env-mediated fusion. Although JR-FL could tolerate substitution of any single CCR5 domain with the corresponding region from CCR2b, SIVmac239 and SIVmac251 could not fuse with cells expressing chimera 5525. Thus, in the context of these chimeras, the second extracellular loop is both necessary and sufficient for fusion cofactor function for these two T-tropic SIVs.
Figure 3.
Differential use of CCR5 by SIV and HIV-1. Cell–cell fusion assays were performed as in Fig. 1, with the indicated env proteins being expressed in HeLa cells and CD4 and the indicated wild-type or chimeric receptor being expressed in QT6 cells. Vertical lines divide chimeras into the following groups: single CCR5 domains in a CCR2b background; single CCR2b domains in a CCR5 background; and multiple domain exchanges.
Although 5222 supported fusion by JR-FL and 2252 supported fusion by SIVmac239 and 251, neither supported fusion as efficiently as wild-type CCR5, suggesting that other regions are required for efficient cofactor function. Therefore, we tested several chimeras containing multiple domain substitutions for their ability to support env-mediated cell–cell fusion. Chimera 2255 supported fusion as efficiently as 2252, indicating that the addition of the third extracellular loop had little effect on cofactor function for SIVmac251 and 239. Chimera 5225 failed to support fusion, again supporting the critical role of the second extracellular loop, and 5252 supported fusion as efficiently as wild-type CCR5. Thus, the combination of the CCR5 amino-terminal domain and second extracellular loop was able to impart efficient cofactor function to CCR2b for JR-FL and T-tropic SIV strains. Chimeras 2552 and 2525 are not processed correctly and so could not be examined (25).
Differential Use of Human CCR5 by SIVs with Different Tropisms.
The results in Fig. 3 show differential use of CCR5 by JR-FL, a representative M-tropic HIV-1 env, and two T-tropic SIV env proteins. We next investigated how CCR5 is used by SIVmac316, an M-tropic variant of SIVmac239. The env protein of SIVmac316 confers M-tropism on SIVmac239 although it differs from SIVmac239 env by only 9 amino acids (38). As shown in Fig. 4, these amino acid changes enabled SIVmac316 env to use 5222 as a fusion cofactor without abolishing its ability to use 2252. In addition, SIVmac316 env could mediate fusion with cells expressing CD4 and 5525 or 5225, albeit less efficiently than JR-FL. These findings indicate that the SIVmac316 env protein could use receptor chimeras containing only the amino-terminal domain of CCR5 and made it less dependent on sequences in the second extracellular loop. Thus, the SIVmac316 env protein closely resembled M-tropic HIV-1 strains in terms of CCR5 usage.
Figure 4.
Differential use of CCR5 by different SIV strains. Cell–cell fusion assays were performed as in Figs. 1 and 3 using SIVmac239 (T-tropic), SIVmac316 (M-tropic), and SIVmac316mut (T-tropic). HIV-1 JR-FL was used as a positive control.
We next examined a mutant form of SIVmac316 env that contains two amino acid changes, P321S and M325I, in the V3 region that renders it T-tropic (39). When tested against the chimeric receptors, we found that this mutant env protein was no longer able to use 5222, 5525, or 5225, indicating that the amino-terminal domain of CCR5 was no longer sufficient to confer cofactor function to CCR2b for this env protein. Rather, the mutant env protein more closely resembled T-tropic SIV stains in its dependence on the second extracellular loop. Thus, specific V3 sequences may be important in determining not only which chemokine receptors are used but also how they are used.
CCR5 Signaling Is Not Required for Cofactor Activity.
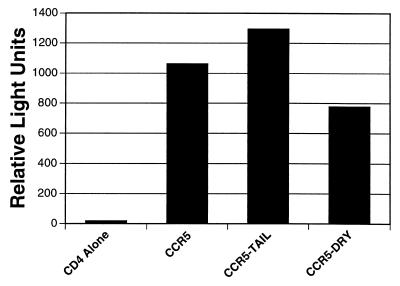
Ligand binding to chemokine receptors results in G protein-coupled signaling events mediated at least in part by sequences in the cytoplasmic domains of the receptor (43). To determine if receptor signaling is required for env-mediated membrane fusion and virus infection, we examined the ability of CCR5 lacking the cytoplasmic domain (CCR5-tail) and CCR5 lacking a conserved DRY motif present in the second intracellular loop to support cell–cell fusion and virus infection. We found that ligand binding to CCR5ΔDRY fails to result in receptor signaling (unpublished work) although ligand binding to CCR5Δtail still results in a calcium flux. We found that both CCR5Δtail and CCR5ΔDRY supported efficient cell–cell fusion by the SIVmac239, 215, 316, and 316mut env proteins (data not shown). In addition, using a luciferase virus reporter system (36, 37), we found that U87-MG cells expressing CD4 and either CCR5Δtail or CCR5ΔDRY supported infection by SIVmac251 as efficiently as wild-type CCR5 (Fig. 5). Thus, G protein-mediated signal transduction is not required for membrane fusion.
Figure 5.
Receptor signaling is not required for virus infection. U87 cells expressing CCR5, CCR5-DRY, or CCR5-tail in conjunction with CD4 were tested for the ability to support virus infection using luciferase reporter viruses. U87-MG cells were transfected with pT4 and either pcDNA3-CCR5, pcDNA3-CKR5Δtail, pcDNA3-CKR5ΔDRY, pcDNA3-CXCR4, or pcDNA3 vector. Cells were infected 24 h posttransfection with 50 ng of BK28 and assayed for luciferase activity 4 days postinfection.
DISCUSSION
The identification of chemokine receptors as cofactors for HIV-1 has important implications for understanding viral tropism, pathogenesis, and the mechanism by which HIV enters cells. In addition, the high degree of protection conferred by a CCR5 polymorphism that renders ≈1% of the Caucasian population effectively CCR5-negative suggests that chemokine receptors may represent important new targets for anti-viral therapies (16–19). Although studying virus–chemokine receptor interactions in vitro has been extremely informative, important questions about the evolution of virus–chemokine receptor interactions during the course of infection in vivo are more difficult to address. SIV represents an important model system for the study of AIDS and potential anti-viral therapies. Through the use of well defined, pathogenic molecular clones, it will be possible to assess the role of chemokine receptor use during virus transmission and to correlate the evolution of virus–chemokine receptor usage with disease progression.
As for HIV-1, CD4 by itself is not sufficient for SIV env-mediated membrane fusion or virus infection (44, 45). Given the similarities between HIV-1 and SIV env proteins, it seemed likely that chemokine receptors also would serve as cofactors for SIV-mediated membrane fusion and virus infection. Indeed, we found that, in the presence of human CD4, CCR5 supported membrane fusion by all SIV strains tested. The use of CCR5 as a cofactor for both M-tropic SIV strains is consistent with it being the major cofactor for M-tropic HIV-1 strains and with it also being used by many syncytium-inducing, primary virus isolates (8). However, the failure of the four T-tropic SIV strains we tested to use CXCR4 was surprising because this is the major cofactor for T-tropic HIV-1 strains (1, 3, 7, 8). The inability of the T-tropic SIV env proteins to use human CXCR4 was not due to a species barrier because rhesus CXCR4 also failed to serve as a fusion cofactor for these isolates. Important to note, rhesus CXCR4 did support fusion by the T-tropic HIV-1 strain BH8, indicating that this construct was expressed on the cell surface and was fully functional. The ability of many SIV strains to infect cells that lack CCR5, such as CEMx174 cells, indicates that simian homologs of other chemokine receptors, or receptors that have not yet been identified, may function as cofactors for these viruses.
Although the discovery of additional coreceptors may help explain how SIV enters CCR5-negative cells, it is not clear why viruses that can use CCR5 fail to replicate in macrophages in an env-dependent manner. SIVmac239, a strictly T-tropic SIV strain, enters macrophages as efficiently as M-tropic SIV isolates yet fails to replicate (41). Furthermore, entry is CD4-dependent and occurs with normal kinetics. Thus, even though the restricted replication of SIVmac239 in macrophages is due to env, it is not due to an entry deficit (41, 42). Our finding that SIVmac239 can efficiently use CCR5 as a fusion cofactor is consistent with its ability to enter macrophages but offers little insight into what is apparently an env-dependent, postentry restriction of virus replication in macrophages. However, we found that two T-tropic SIV strains used CCR5 in a manner that is distinct from that seen with a closely related M-tropic SIV isolate. We have shown previously that the amino-terminal domain of CCR5 is the only domain that, when introduced into CCR2b, is able to confer HIV-1 cofactor function (25), a finding that recently has been confirmed (26). Analysis of additional chimeric and mutant receptors identified seven that could function for either dual-tropic or M-tropic HIV-1 strains but not both, indicating that different virus strains can exhibit diverse patterns of CCR5 usage.
Our results showed distinct differences in CCR5 use by T- and M-tropic SIV strains. The M-tropic strain SIVmac316 more closely resembled the M-tropic HIV-1 strains JR-FL, SF162, and ADA in terms of chimeric receptor usage, including the ability to use chimera 5222. By contrast, the T-tropic strains SIVmac239 and SIVmac251 required the presence of the second extracellular loop of CCR5 and exhibited a decreased dependence on the amino-terminal domain. These differences result from a relatively small number of amino acid differences in env. SIVmac316 differs from SIVmac239 at six positions in gp120 (none in the V3 region) and at three in gp41, with the changes in gp120 being largely responsible for macrophage tropism (38). Like HIV-1, interactions between SIV and CCR5 are likely to be complex, involving multiple domains of CCR5 and probably both V3 loop and non-V3 loop domains in env. Although differential use of CCR5 by SIVmac316 is not the result of changes in the V3 region of env, substitutions in this domain have been shown to be associated with altered SIV cell tropism (39, 46). Indeed, we found that two amino acid changes in the V3 region (P321S and M325I) of SIVmac316 mut that result in drastically reduced replication in macrophages caused this env to more closely resemble T-tropic SIV strains in terms of CCR5 usage. Most strikingly, this mutant lost the ability to use chimera 5222. Thus, relatively subtle changes in the SIV V3 domain can dramatically alter how a chemokine receptor is used.
These and previous results (25) clearly show differential use of CCR5 by different virus strains. The critical question is whether these differences have consequences for virus infection. All of the SIV strains studied here underwent membrane fusion with cells expressing CCR5 with similar efficiency, which is consistent with the efficient entry of both SIVmac239 and SIVmac316 into macrophages. Likewise, the dual-tropic HIV-1 strain 89.6 fuses with cells expressing CCR5 as well as CCR5-restricted viruses such as JR-FL. Thus, at least with the assay used in our studies, differential CCR5 usage does not seem to result in altered membrane fusion efficiencies. An intriguing possibility, suggested by Mori et al. (41), is that the way in which env protein interacts with the cell surface can directly influence postentry events, perhaps by inducing receptor signaling. Although G protein-mediated CCR5 signal transduction is clearly not required for the membrane fusion reaction, it could influence postentry events that are critical for a productive SIV infection of macrophages in certain contexts. Indeed, a variety of factors have been shown to regulate virus expression in macrophages (31). Although env has been shown to interact with CCR5 (23, 24), it is not yet known whether this interaction results in any type of receptor signaling.
M-tropic strains of HIV-1 use CCR5 as a fusion cofactor and are responsible for sexual transmission. The finding that individuals who lack CCR5 are highly resistant to virus infection by both M- and T-tropic viruses suggests that CXCR4-restricted viruses are inefficient at establishing an infection in a naive host (16–19). In contrast, T-tropic strains of SIV can readily establish an infection in which M-tropic viruses arise over time. This may reflect the ability of T-tropic SIV strains to use CCR5, analogous to M-tropic strains of HIV-1. However, the identification of additional SIV cofactors and studies on chemokine receptor distribution will be required to more fully understand the role of chemokine receptor use in SIV transmission and disease progression. In addition, it will be important to determine if additional primary, T-tropic SIV isolates also use CCR5, particularly isolates that have not been passaged in transformed cell lines. Finally, our results raise the possibility that viral tropism may be determined in part by the cofactor domains that interact with env (gp120), as well as by the repertoire of cofactors used by a given virus strain.
Acknowledgments
We thank Ron Desrosiers for kindly providing us with clones for the SIVmac239, 251, 316, and 316mut env proteins, Ned Landau for reagents and expert advice on the luciferase virus system, and Joe Rucker for helpful discussions and assistance with the figures. A.L.E. was supported by the National Institutes of Health Medical Scientist Training Program, and B.J.D. and K.M. were supported by Howard Hughes Medical Institute Predoctoral Fellowships. This work was supported by National Institutes of Health Grants AI-35383, AI-38225, and AI-40880 to R.W.D.
ABBREVIATIONS
- HIV-1
HIV type 1
- envelope
env
- M-tropic
macrophage tropic
- T-tropic
T cell tropic
- SIV
simian immunodeficiency virus
References
- 1.Feng Y, Broder C C, Kennedy P E, Berger E A. Science. 1996;272:872–877. [Google Scholar]
- 2.Alkhatib G, Combadiere C, Broder C C, Feng Y, Kennedy P E, Murphy P M, Berger E A. Science. 1996;272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 3.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath P D, Wu L, Mackay C R, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. Cell. 1996;85:1135–1148. doi: 10.1016/s0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 4.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Marzio P D, Marmon S, Sutton R E, Hill C M, Davis C B, Peiper S C, Schall T J, Littman D R, Landau N R. Nature (London) 1996;381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 5.Doranz B J, Rucker J, Yi Y, Smyth R J, Samson M, Peiper S C, Parmentier M, Collman R G, Doms R W. Cell. 1996;85:1149–1158. doi: 10.1016/s0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 6.Dragic T, Litwin V, Allaway G P, Martin S R, Huang Y, Nagashima K A, Cayanan C, Maddon P J, Koup R A, Moore J P, Paxton W A. Nature (London) 1996;381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 7.Berson J F, Long D, Doranz B J, Rucker J, Jirik F R, Doms R W. J Virol. 1996;70:6288–6295. doi: 10.1128/jvi.70.9.6288-6295.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Simmons G, Wilkinson D, Reeves J D, Dittman M T, Beddows S, Weber J, Carnegis G, Gesselberger U, Gray P W, Weiss R A, Clapham P R. J Virol. 1996;70:8355–8360. doi: 10.1128/jvi.70.12.8355-8360.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tersmette, M., deGoede, R. E. Y., Ai, B. J. M., Winkel, I. N., Gruters, R. A., Cuypers, H. T., Huisman, H. G. & Miedema, F. (1988) J. Virol. 62. [DOI] [PMC free article] [PubMed]
- 10.Tersmette M, Gruters R, Wolf F d, Goede R E Y d, Lange J M A, Schellekens P T A, Goudsmit J, Huisman H G, Miedema F. J Virol. 1989;63:2118–2125. doi: 10.1128/jvi.63.5.2118-2125.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Conner R I, Mohri H, Cao Y, Ho D D. J Virol. 1993;67:1772–1777. doi: 10.1128/jvi.67.4.1772-1777.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Roos M T L, Lange J M A, Goede R E Y d, Coutinho R A, Schellekens P T A, Miedema F, Tersmette M. J Infect Dis. 1992;165:427–432. doi: 10.1093/infdis/165.3.427. [DOI] [PubMed] [Google Scholar]
- 13.Zho T, Mo H, Wang N, Nam D S, Cao Y, Koup R A, Ho D D. Science. 1993;261:1179–1181. doi: 10.1126/science.8356453. [DOI] [PubMed] [Google Scholar]
- 14.Schuitemaker H, Kootstra N A, Goede R E Y D, Wolf F d, Miedema F, Tersmette M. J Virol. 1991;65:356–363. doi: 10.1128/jvi.65.1.356-363.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schuitemaker H, Koot M, Koostra N A, Dercksen M W, Goede R E Y d, Steenwijk R P v, Lange J M A, Schattenkerk J K M E, Miedema F, Tersmette M. J Virol. 1992;66:1354–1360. doi: 10.1128/jvi.66.3.1354-1360.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Samson M, Libert F, Doranz B J, Rucker J, Liesnard C, et al. Nature (London) 1996;382:722–725. doi: 10.1038/382722a0. [DOI] [PubMed] [Google Scholar]
- 17.Liu R, Paxton W A, Choe S, Ceradini D, Martin S R, Horuk R, MacDonald M E, Stuhlmann H, Koup R A, Landau N R. Cell. 1996;86:367–377. doi: 10.1016/s0092-8674(00)80110-5. [DOI] [PubMed] [Google Scholar]
- 18.Huang Y, Paxton W A, Wolinsky S M, Neumann A U, Zhang L, He T, Kang S, Ceradini D, Jin Z, Yazdanbakhsh K, Kuntsman K, Erickson D, Dragon E, Landau N R, Phair J, Ho D D, Koup R A. Nat Med. 1996;2:1240–1243. doi: 10.1038/nm1196-1240. [DOI] [PubMed] [Google Scholar]
- 19.Dean M, Carrington M, Winkler C, Huttley G A, Smith M W, Allikmets R, Goedert J J, Buchbinder S P, Vittinghoff E, Gomperts E, Donfield S, Vlahov D, Kaslow R, Saah A, Rinaldo C, Detels R, O’Brien S J. Science. 1996;273:1856–1862. doi: 10.1126/science.273.5283.1856. [DOI] [PubMed] [Google Scholar]
- 20.Cocchi F, DeVico A L, Garzino-Demo A, Arya S K, Gallo R C, Lusso P. Science. 1995;270:1811–1815. doi: 10.1126/science.270.5243.1811. [DOI] [PubMed] [Google Scholar]
- 21.Paxton W A, Martin S R, Tse D, O’Brien T R, Skurnick J, VanDevanter N L, Padian N, Braun J F, Kotler D P, Wolinsky S M, Koup R A. Nat Med. 1996;2:412–417. doi: 10.1038/nm0496-412. [DOI] [PubMed] [Google Scholar]
- 22.Lapham C K, Ouyang J, Chandrasekhar B, Nguyen N Y, Dimitrov D S, Golding H. Science. 1996;274:602–605. doi: 10.1126/science.274.5287.602. [DOI] [PubMed] [Google Scholar]
- 23.Wu L, Gerard N P, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso A A, Desjardin E, Newman W, Gerard C, Sodroski J. Nature (London) 1996;384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 24.Trkola A, Dragic T, Arthos J, Binley J M, Olson W C, Allaway G P, Cheng-Mayer C, Robinson J, Maddon P J, Moore J P. Nature (London) 1996;384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 25.Rucker J, Samson M, Doranz B J, Libert F, Berson J, Yi Y, Collman R G, Vassart G, Broder C C, Doms R W, Parmentier M. Cell. 1996;87:437–446. doi: 10.1016/s0092-8674(00)81364-1. [DOI] [PubMed] [Google Scholar]
- 26.Atchison R E, Gosling J, Monteclaro F S, Franci C, Digilio L, Charo I F, Goldsmith M. Science. 1996;274:1924–1926. doi: 10.1126/science.274.5294.1924. [DOI] [PubMed] [Google Scholar]
- 27.Chesebro B, Nishis J, Perryman S, Cann A, O’Brien W, Chen I, Wehrly K. J Virol. 1991;65:57832–5789. doi: 10.1128/jvi.65.11.5782-5789.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cheng-Mayer C, Quiroga M, Tung J W, Dina D, Levy J A. J Virol. 1990;64:4390–4398. doi: 10.1128/jvi.64.9.4390-4398.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hwang S S, Boyle T J, Lyerly H K, Cullen B R. Science. 1991;253:71–74. doi: 10.1126/science.1905842. [DOI] [PubMed] [Google Scholar]
- 30.Amedee A M, Lacour N, Gierman J L, Martin L N, Clements J E, Jr, R B, Harrison R M, Murphey-Corb M. J Virol. 1995;69:7982–7990. doi: 10.1128/jvi.69.12.7982-7990.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fauci A. Nature (London) 1996;384:529–534. doi: 10.1038/384529a0. [DOI] [PubMed] [Google Scholar]
- 32.Anderson M G, Hauer D, Sharma D P, Joag S V, Narayan O, Zink M C, Clements J E. Virology. 1993;195:616–626. doi: 10.1006/viro.1993.1413. [DOI] [PubMed] [Google Scholar]
- 33.Broder C C, Berger E A. Proc Natl Acad Sci USA. 1995;92:9004–9008. doi: 10.1073/pnas.92.19.9004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alexander W A, Moss B, Fuerst T R. J Virol. 1992;66:2934–2942. doi: 10.1128/jvi.66.5.2934-2942.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nussbaum O, Broder C C, Berger E A. J Virol. 1994;68:5411–5422. doi: 10.1128/jvi.68.9.5411-5422.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen B K, Saksela K, Andino R, Baltimore D. J Virol. 1994;68:654–660. doi: 10.1128/jvi.68.2.654-660.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Conner R I, Chen B K, Choe S, Landau N R. Virology. 1995;206:935–944. doi: 10.1006/viro.1995.1016. [DOI] [PubMed] [Google Scholar]
- 38.Mori K, Ringler D J, Kodama T, Desrosiers R C. J Virol. 1992;66:2067–2075. doi: 10.1128/jvi.66.4.2067-2075.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kirchhoff F, Mori K, Desrosiers R C. J Virol. 1994;68:3682–3692. doi: 10.1128/jvi.68.6.3682-3692.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clements J E, Zink M C. Clin Microbiol Rev. 1996;9:100–117. doi: 10.1128/cmr.9.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Mori K, Ringler D J, Desrosiers R C. J Virol. 1993;67:2807–2814. doi: 10.1128/jvi.67.5.2807-2814.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stephens E B, McClure H M, Narayan O. Virology. 1995;206:535–544. doi: 10.1016/s0042-6822(95)80070-0. [DOI] [PubMed] [Google Scholar]
- 43.Probst W C, Snyder L A, Schuster D I, Brosius J, Sealfon S C. DNA Cell Biol. 1996;11:1–20. doi: 10.1089/dna.1992.11.1. [DOI] [PubMed] [Google Scholar]
- 44.McKnight A, Clapham P R, Weiss R A. Virology. 1994;201:8–18. doi: 10.1006/viro.1994.1260. [DOI] [PubMed] [Google Scholar]
- 45.Clapham P R, Blanc D, Weiss R A. Virology. 1991;181:703–715. doi: 10.1016/0042-6822(91)90904-P. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kodama T, Mori K, Kawahara T, Ringler D J, Desrosiers R C. J Virol. 1993;67:6522–6534. doi: 10.1128/jvi.67.11.6522-6534.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]