Abstract
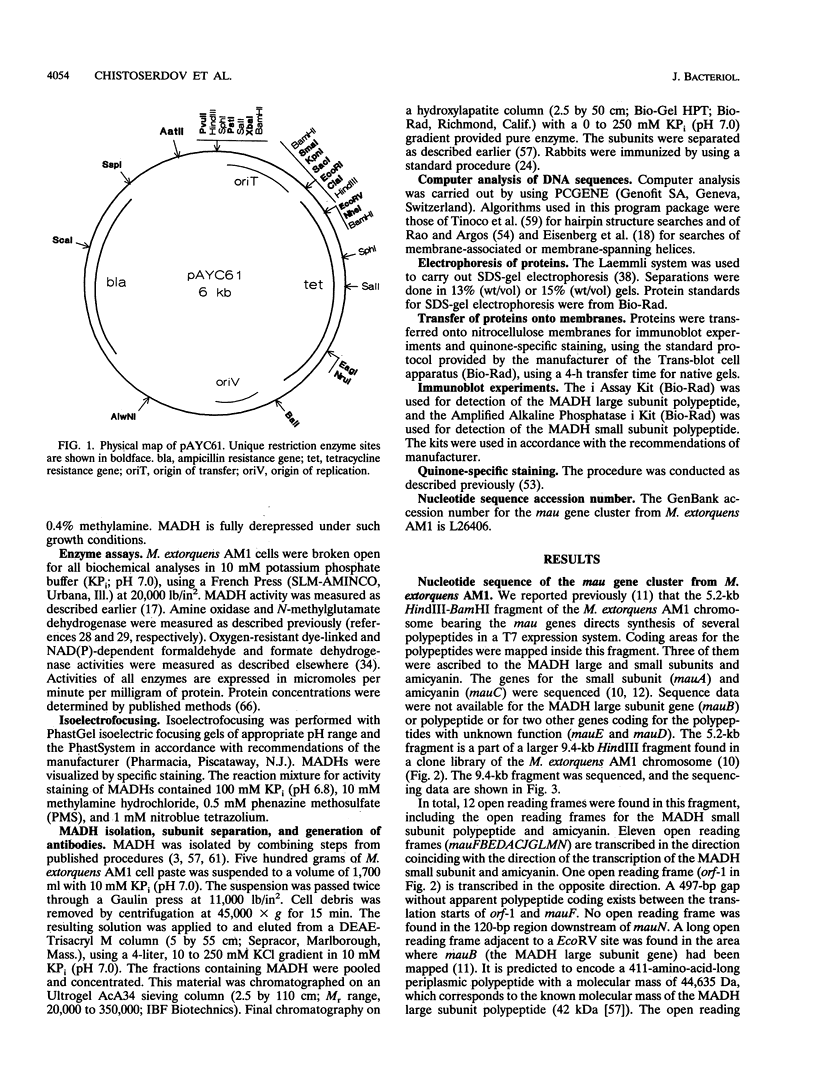
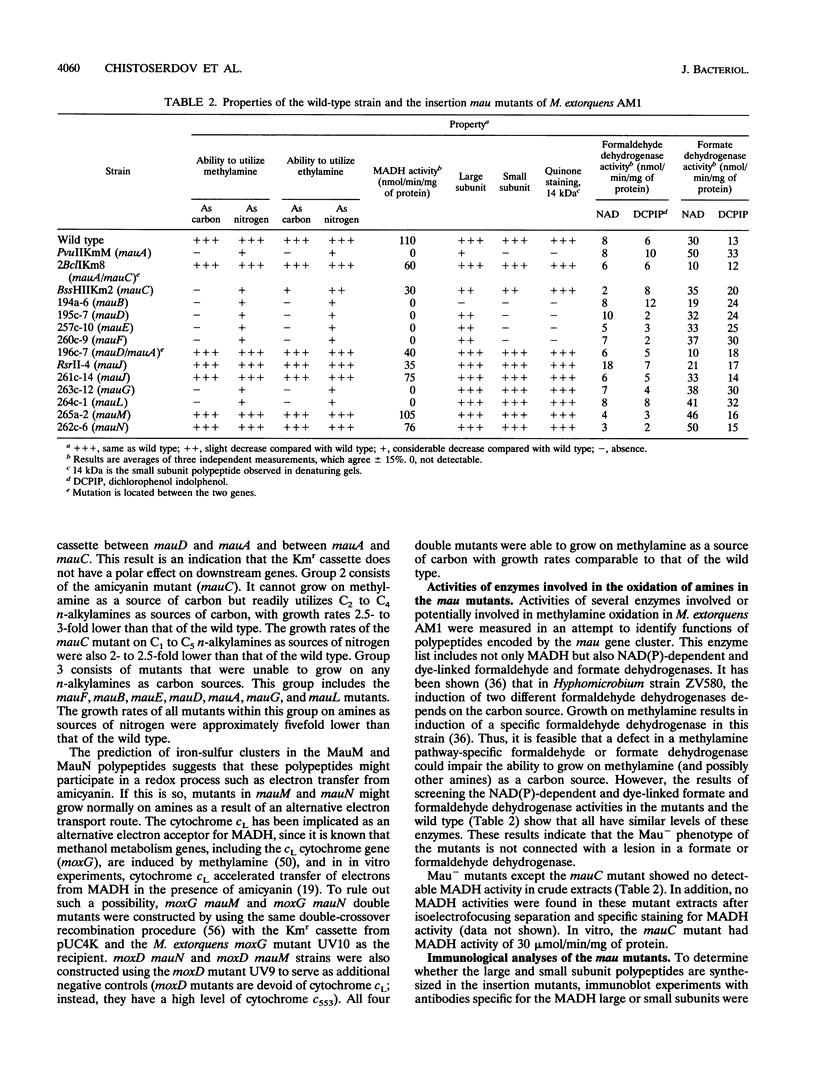
The nucleotide sequence of the methylamine utilization (mau) gene region from Methylobacterium extorquens AM1 was determined. Open reading frames for 11 genes (mauFBEDACJGLMN) were found, all transcribed in the same orientation. The mauB, mauA, and mauC genes encode the periplasmic methylamine dehydrogenase (MADH) large and small subunit polypeptides and amicyanin, respectively. The products of mauD, mauG, mauL, and mauM were also predicted to be periplasmic. The products of mauF, mauE, and mauN were predicted to be membrane associated. The mauJ product is the only polypeptide encoded by the mau gene cluster which is predicted to be cytoplasmic. Computer analysis showed that the MauG polypeptide contains two putative heme binding sites and that the MauM and MauN polypeptides have four and two FeS cluster signatures, respectively. Mutants generated by insertions in mauF, mauB, mauE, mauD, mauA, mauG, and mauL were not able to grow on methylamine or any other primary amine as carbon sources, while a mutant generated from an insertion in mauC was not able to utilize methylamine as a source of carbon but utilized C2 to C4 n-alkylamines as carbon sources. Insertion mutations in mauJ, mauM, and mauN did not impair the ability of the mutants to utilize primary n-alkylamines as carbon sources. All mau mutants were able to utilize methylamine as a nitrogen source, implying the existence of an alternative (methyl)amine oxidation system, and a low activity of N-methylglutamate dehydrogenase was detected. The mauD, mauE, and mauF mutants were found to lack the MADH small subunit polypeptide and have a decreased amount of the MADH large subunit polypeptide. In the mauG and mauL mutants, the MADH large and small subunit polypeptides were present at wild-type levels, although the MADHs in these strains were not functional. In addition, MauG has sequence similarity to cytochrome c peroxidase from Pseudomonas sp. The mauA, mauD, and mauE genes from Paracoccus denitrificans and the mauD and mauG genes from Methylophilus methylotrophus W3A1 were able to complement corresponding mutants of M. extorquens AM1, confirming their functional equivalence. Comparison of amino acid sequences of polypeptides encoded by mau genes from M. extorquens AM1, P. denitrificans, and Thiobacillus versutus shows that they have considerable similarity.
Full text
PDF











Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Boulton C. A., Large P. J. Properties of Pseudomonas AM1 primary-amine dehydrogenase immobilized on agarose. Biochim Biophys Acta. 1979 Sep 12;570(1):22–30. doi: 10.1016/0005-2744(79)90197-9. [DOI] [PubMed] [Google Scholar]
- Chandrasekar R., Klapper M. H. Methylamine dehydrogenase and cytochrome c552 from the bacterium W3A1. J Biol Chem. 1986 Mar 15;261(8):3616–3619. [PubMed] [Google Scholar]
- Chistoserdov A. Y., Lidstrom M. E. The small-subunit polypeptide of methylamine dehydrogenase from Methylobacterium extorquens AM1 has an unusual leader sequence. J Bacteriol. 1991 Sep;173(18):5909–5913. doi: 10.1128/jb.173.18.5909-5913.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdov A. Y., McIntire W. S., Mathews F. S., Lidstrom M. E. Organization of the methylamine utilization (mau) genes in Methylophilus methylotrophus W3A1-NS. J Bacteriol. 1994 Jul;176(13):4073–4080. doi: 10.1128/jb.176.13.4073-4080.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdov A. Y., Tsygankov Y. D., Lidstrom M. E. Genetic organization of methylamine utilization genes from Methylobacterium extorquens AM1. J Bacteriol. 1991 Sep;173(18):5901–5908. doi: 10.1128/jb.173.18.5901-5908.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chistoserdov A. Y., Tsygankov Y. D., Lidstrom M. E. Nucleotide sequence of the amicyanin gene from Methylobacterium extorquens AM1. DNA Seq. 1991;2(1):53–55. doi: 10.3109/10425179109008439. [DOI] [PubMed] [Google Scholar]
- Ditta G., Schmidhauser T., Yakobson E., Lu P., Liang X. W., Finlay D. R., Guiney D., Helinski D. R. Plasmids related to the broad host range vector, pRK290, useful for gene cloning and for monitoring gene expression. Plasmid. 1985 Mar;13(2):149–153. doi: 10.1016/0147-619x(85)90068-x. [DOI] [PubMed] [Google Scholar]
- Eady R. R., Large P. J. Purification and properties of an amine dehydrogenase from Pseudomonas AM1 and its role in growth on methylamine. Biochem J. 1968 Jan;106(1):245–255. doi: 10.1042/bj1060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenberg D., Schwarz E., Komaromy M., Wall R. Analysis of membrane and surface protein sequences with the hydrophobic moment plot. J Mol Biol. 1984 Oct 15;179(1):125–142. doi: 10.1016/0022-2836(84)90309-7. [DOI] [PubMed] [Google Scholar]
- Fukumori Y., Yamanaka T. The methylamine oxidizing system of Pseudomonas AM1 reconstituted with purified components. J Biochem. 1987 Feb;101(2):441–445. doi: 10.1093/oxfordjournals.jbchem.a121929. [DOI] [PubMed] [Google Scholar]
- Fulton G. L., Nunn D. N., Lidstrom M. E. Molecular cloning of a malyl coenzyme A lyase gene from Pseudomonas sp. strain AM1, a facultative methylotroph. J Bacteriol. 1984 Nov;160(2):718–723. doi: 10.1128/jb.160.2.718-723.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gennity J., Goldstein J., Inouye M. Signal peptide mutants of Escherichia coli. J Bioenerg Biomembr. 1990 Jun;22(3):233–269. doi: 10.1007/BF00763167. [DOI] [PubMed] [Google Scholar]
- Guerlesquin F., Bruschi M., Bovier-Lapierre G., Bonicel J., Couchoud P. Primary structure of the two (4 Fe-4 S) clusters ferredoxin from Desulfovibrio desulfuricans (strain Norway 4). Biochimie. 1983 Jan;65(1):43–47. doi: 10.1016/s0300-9084(83)80027-3. [DOI] [PubMed] [Google Scholar]
- Harms N., Reijnders W. N., Anazawa H., van der Palen C. J., van Spanning R. J., Oltmann L. F., Stouthamer A. H. Identification of a two-component regulatory system controlling methanol dehydrogenase synthesis in Paracoccus denitrificans. Mol Microbiol. 1993 May;8(3):457–470. doi: 10.1111/j.1365-2958.1993.tb01590.x. [DOI] [PubMed] [Google Scholar]
- Hausinger R. P., Moura I., Moura J. J., Xavier A. V., Santos M. H., LeGall J., Howard J. B. Amino acid sequence of a 3Fe:3S ferredoxin from the "archaebacterium" Methanosarcina barkeri (DSM 800). J Biol Chem. 1982 Dec 10;257(23):14192–14197. [PubMed] [Google Scholar]
- Haywood G. W., Large P. J. Microbial oxidation of amines. Distribution, purification and properties of two primary-amine oxidases from the yeast Candida boidinii grown on amines as sole nitrogen source. Biochem J. 1981 Oct 1;199(1):187–201. doi: 10.1042/bj1990187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hersh L. B., Peterson J. A., Thompson A. A. An N-methyl glutamate dehydrogenase from Pseudomonas M.A. Arch Biochem Biophys. 1971 Jul;145(1):115–120. doi: 10.1016/0003-9861(71)90016-6. [DOI] [PubMed] [Google Scholar]
- Huitema F., van Beeumen J., van Driessche G., Duine J. A., Canters G. W. Cloning and sequencing of the gene coding for the large subunit of methylamine dehydrogenase from Thiobacillus versutus. J Bacteriol. 1993 Oct;175(19):6254–6259. doi: 10.1128/jb.175.19.6254-6259.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Husain M., Davidson V. L. An inducible periplasmic blue copper protein from Paracoccus denitrificans. Purification, properties, and physiological role. J Biol Chem. 1985 Nov 25;260(27):14626–14629. [PubMed] [Google Scholar]
- Husain M., Davidson V. L. Purification and properties of methylamine dehydrogenase from Paracoccus denitrificans. J Bacteriol. 1987 Apr;169(4):1712–1717. doi: 10.1128/jb.169.4.1712-1717.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson P. A., Quayle J. R. Microbial growth on C-1 compounds. 6. Oxidation of methanol, formaldehyde and formate by methanol-grown Pseudomonas AM-1. Biochem J. 1964 Nov;93(2):281–290. doi: 10.1042/bj0930281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenney W. C., McIntire W. Characterization of methylamine dehydrogenase from bacterium W3A1. Interaction with reductants and amino-containing compounds. Biochemistry. 1983 Aug 2;22(16):3858–3868. doi: 10.1021/bi00285a022. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Levering P. R., van Dijken J. P., Veenhius M., Harder W. Arthrobacter P1, a fast growing versatile methylotroph with amine oxidase as a key enzyme in the metabolism of methylated amines. Arch Microbiol. 1981 Mar;129(1):72–80. doi: 10.1007/BF00417184. [DOI] [PubMed] [Google Scholar]
- McIntire W. S., Wemmer D. E., Chistoserdov A., Lidstrom M. E. A new cofactor in a prokaryotic enzyme: tryptophan tryptophylquinone as the redox prosthetic group in methylamine dehydrogenase. Science. 1991 May 10;252(5007):817–824. doi: 10.1126/science.2028257. [DOI] [PubMed] [Google Scholar]
- Meinkoth J., Wahl G. Hybridization of nucleic acids immobilized on solid supports. Anal Biochem. 1984 May 1;138(2):267–284. doi: 10.1016/0003-2697(84)90808-x. [DOI] [PubMed] [Google Scholar]
- Mohana Rao J. K., Argos P. A conformational preference parameter to predict helices in integral membrane proteins. Biochim Biophys Acta. 1986 Jan 30;869(2):197–214. doi: 10.1016/0167-4838(86)90295-5. [DOI] [PubMed] [Google Scholar]
- Morris C. J., Lidstrom M. E. Cloning of a methanol-inducible moxF promoter and its analysis in moxB mutants of Methylobacterium extorquens AM1rif. J Bacteriol. 1992 Jul;174(13):4444–4449. doi: 10.1128/jb.174.13.4444-4449.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nunn D. N., Lidstrom M. E. Isolation and complementation analysis of 10 methanol oxidation mutant classes and identification of the methanol dehydrogenase structural gene of Methylobacterium sp. strain AM1. J Bacteriol. 1986 May;166(2):581–590. doi: 10.1128/jb.166.2.581-590.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott S., Jones S., Cooper R. A. 2-Phenylethylamine catabolism by Escherichia coli K12. J Gen Microbiol. 1987 Feb;133(2):347–351. doi: 10.1099/00221287-133-2-347. [DOI] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Rönnberg M., Kalkkinen N., Ellfolk N. The primary structure of Pseudomonas cytochrome c peroxidase. FEBS Lett. 1989 Jul 3;250(2):175–178. doi: 10.1016/0014-5793(89)80714-8. [DOI] [PubMed] [Google Scholar]
- Shirai S., Matsumoto T., Tobari J. Methylamine dehydrogenase of Pseudomonas AM1. A subunit enzyme. J Biochem. 1978 Jun;83(6):1599–1607. doi: 10.1093/oxfordjournals.jbchem.a132071. [DOI] [PubMed] [Google Scholar]
- Simon R. High frequency mobilization of gram-negative bacterial replicons by the in vitro constructed Tn5-Mob transposon. Mol Gen Genet. 1984;196(3):413–420. doi: 10.1007/BF00436188. [DOI] [PubMed] [Google Scholar]
- Tinoco I., Jr, Borer P. N., Dengler B., Levin M. D., Uhlenbeck O. C., Crothers D. M., Bralla J. Improved estimation of secondary structure in ribonucleic acids. Nat New Biol. 1973 Nov 14;246(150):40–41. doi: 10.1038/newbio246040a0. [DOI] [PubMed] [Google Scholar]
- Tobari J., Harada Y. Amicyanin: an electron acceptor of methylamine dehydrogenase. Biochem Biophys Res Commun. 1981 Jul 30;101(2):502–508. doi: 10.1016/0006-291x(81)91288-2. [DOI] [PubMed] [Google Scholar]
- Ubbink M., van Kleef M. A., Kleinjan D. J., Hoitink C. W., Huitema F., Beintema J. J., Duine J. A., Canters G. W. Cloning, sequencing and expression studies of the genes encoding amicyanin and the beta-subunit of methylamine dehydrogenase from Thiobacillus versutus. Eur J Biochem. 1991 Dec 18;202(3):1003–1012. doi: 10.1111/j.1432-1033.1991.tb16462.x. [DOI] [PubMed] [Google Scholar]
- Wagner C., Quayle J. R. Carbon assimilation pathways during growth of Pseudomonas AM1 on methylamine and Pseudomonas MS on methylamine and trimethylsulphonium salts. J Gen Microbiol. 1972 Oct;72(3):485–491. doi: 10.1099/00221287-72-3-485. [DOI] [PubMed] [Google Scholar]
- Whitaker J. R., Granum P. E. An absolute method for protein determination based on difference in absorbance at 235 and 280 nm. Anal Biochem. 1980 Nov 15;109(1):156–159. doi: 10.1016/0003-2697(80)90024-x. [DOI] [PubMed] [Google Scholar]
- d'Ischia M., Napolitano A., Prota G. Peroxidase as an alternative to tyrosinase in the oxidative polymerization of 5,6-dihydroxyindoles to melanin(s). Biochim Biophys Acta. 1991 Mar 4;1073(2):423–430. doi: 10.1016/0304-4165(91)90152-7. [DOI] [PubMed] [Google Scholar]
- de Moura Gallo C. V., Vassetzky Y. S., Huesca M., Scherrer K. A transcription-dependent DNase I-hypersensitive site in a far upstream segment of the chicken alpha-globin gene domain coincides with a matrix attachment region. Biochem Biophys Res Commun. 1992 May 15;184(3):1226–1234. doi: 10.1016/s0006-291x(05)80013-0. [DOI] [PubMed] [Google Scholar]
- van Houwelingen T., Canters G. W., Stobbelaar G., Duine J. A., Frank J., Jr, Tsugita A. Isolation and characterization of a blue copper protein from Thiobacillus versutus. Eur J Biochem. 1985 Nov 15;153(1):75–80. doi: 10.1111/j.1432-1033.1985.tb09268.x. [DOI] [PubMed] [Google Scholar]
- van Spanning R. J., Wansell C. W., Reijnders W. N., Oltmann L. F., Stouthamer A. H. Mutagenesis of the gene encoding amicyanin of Paracoccus denitrificans and the resultant effect on methylamine oxidation. FEBS Lett. 1990 Nov 26;275(1-2):217–220. doi: 10.1016/0014-5793(90)81475-4. [DOI] [PubMed] [Google Scholar]