Abstract
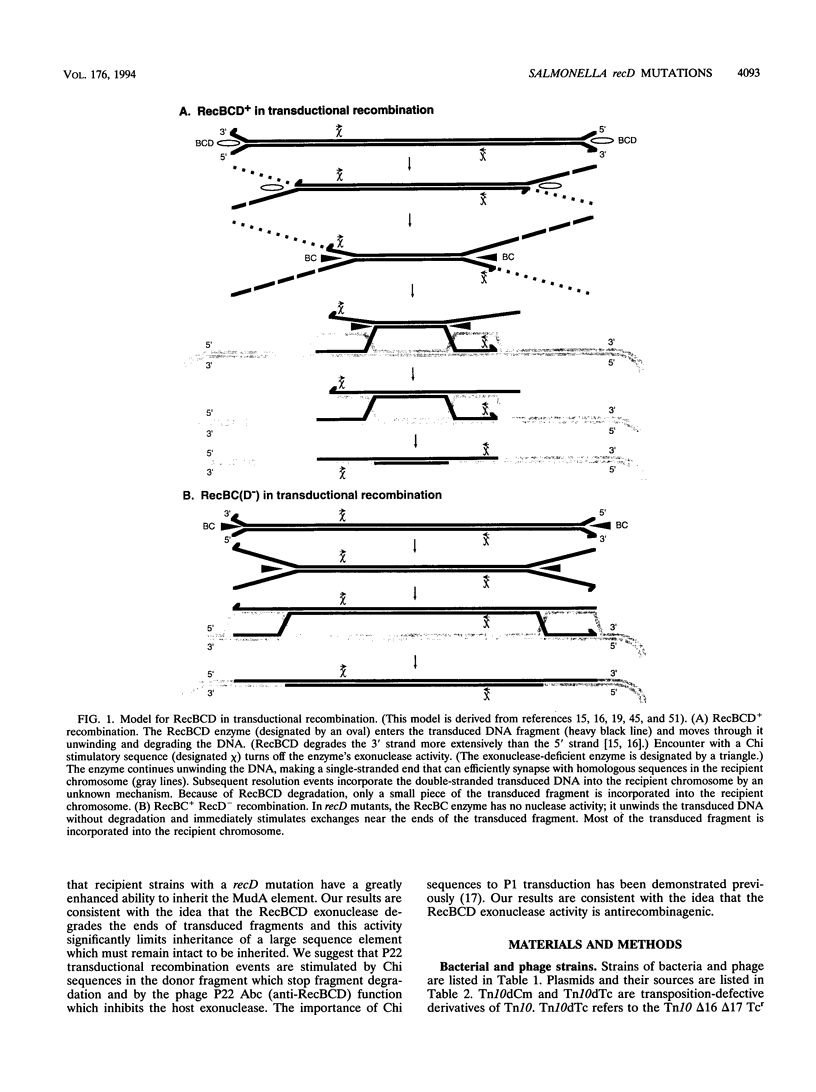
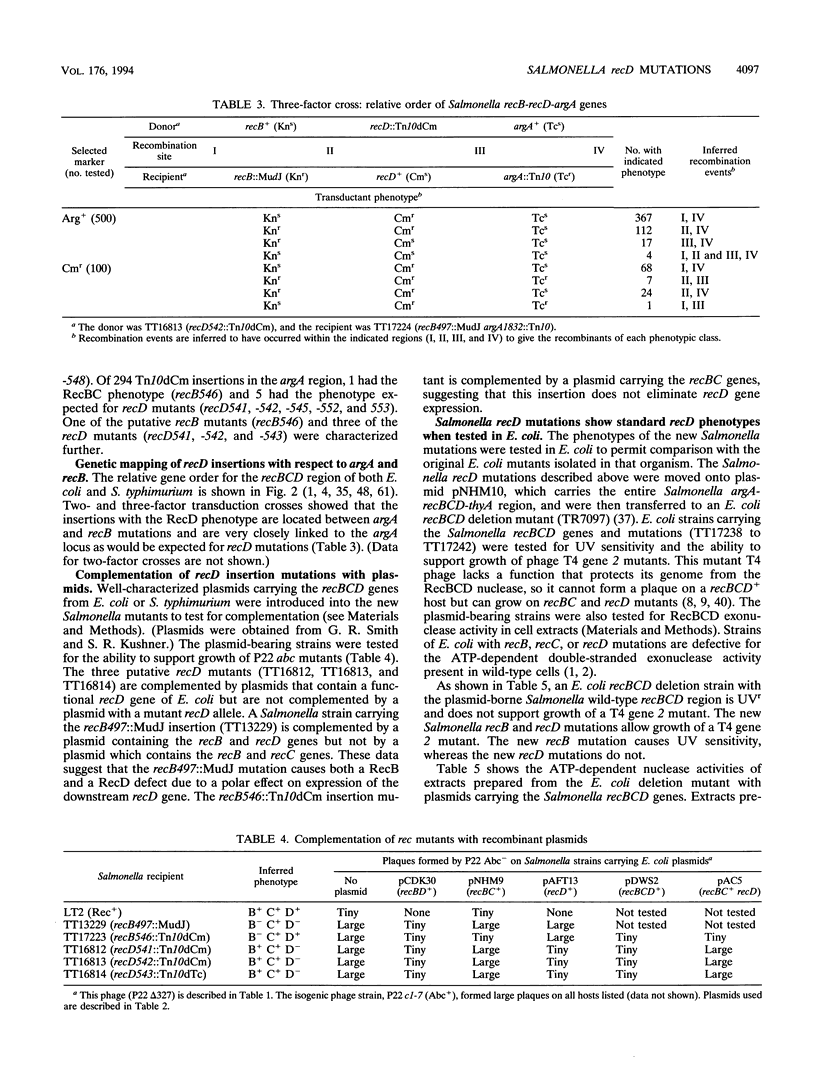
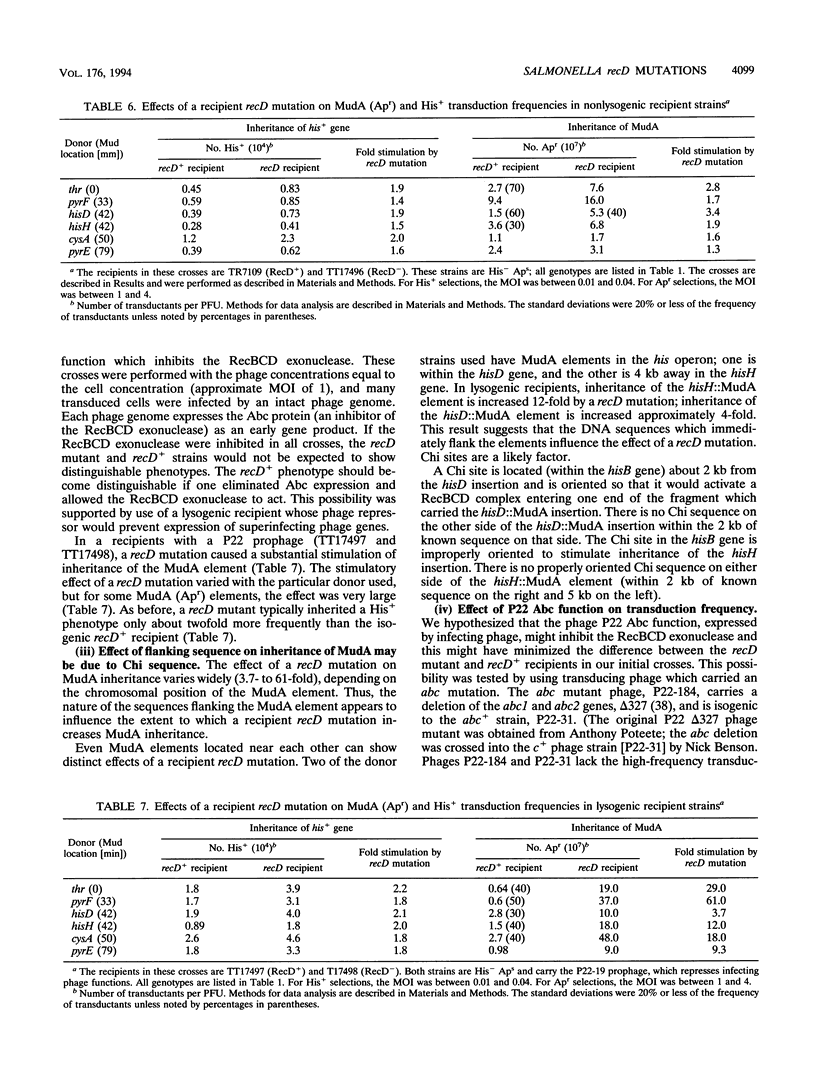
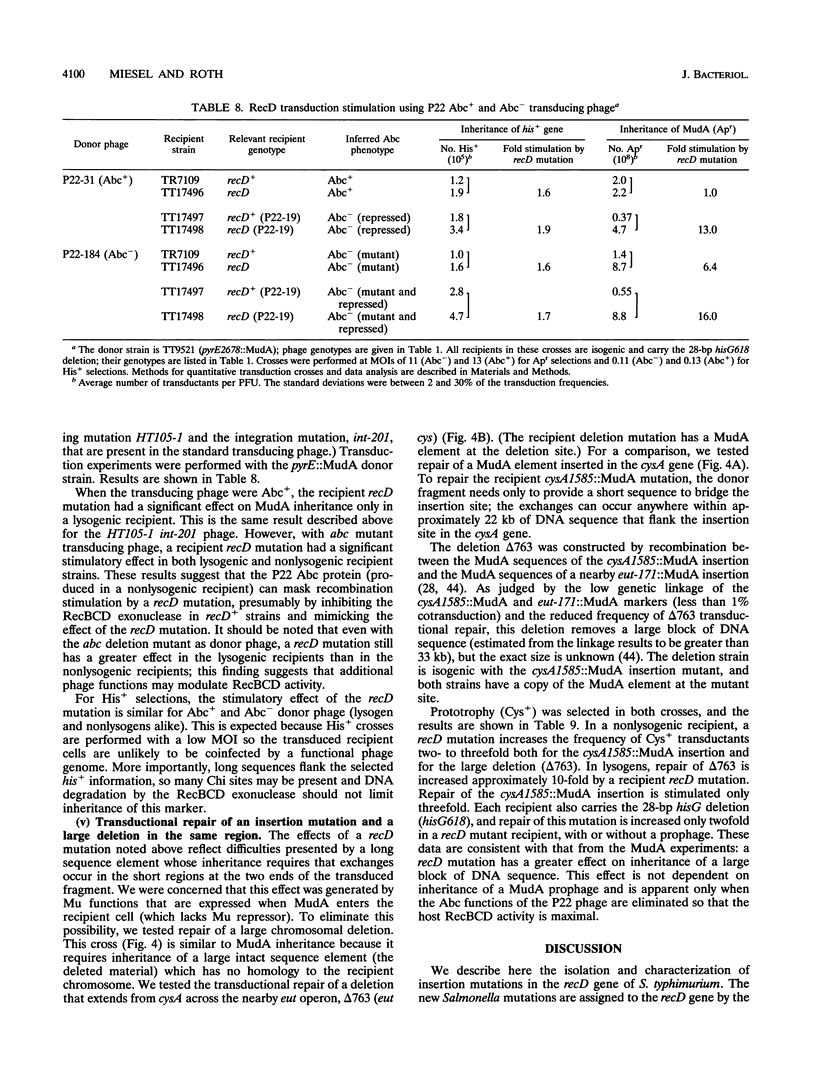
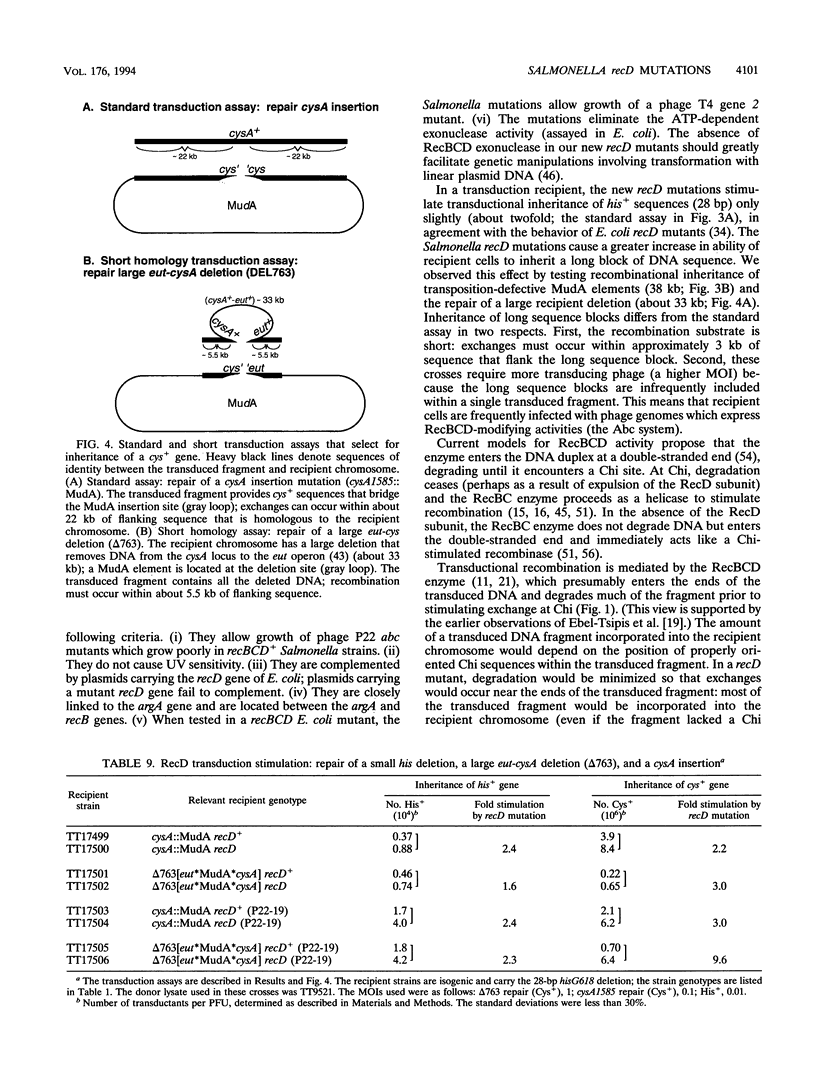
We have identified recD mutants of Salmonella typhimurium by their ability to support growth of phage P22 abc (anti-RecBCD) mutants, whose growth is prevented by normal host RecBCD function. As in Escherichia coli, the recD gene of S. typhimurium lies between the recB and argA genes at min 61 of the genetic map. Plasmids carrying the Salmonella recBCD+ genes restore ATP-dependent exonuclease V activity to an E. coli recBCD deletion mutant. The new Salmonella recD mutations (placed on this plasmid) eliminate the exonuclease activity and enable the plasmid-bearing E. coli deletion mutant to support growth of phage T4 gene 2 mutants. The Salmonella recD mutations caused a 3- to 61-fold increase in the ability of a recipient strain to inherit (by transduction) a large inserted element (MudA prophage; 38 kb). In this cross, recombination events must occur in the short (3-kb) sequences that flank the element in the 44-kb transduced fragment. The effect of the recD mutation depends on the nature of the flanking sequences and is likely to be greatest when those sequences lack a Chi site. The recD mutation appears to minimize fragment degradation and/or cause RecBC-dependent recombination events to occur closer to the ends of the transduced fragment. The effect of a recipient recD mutation was eliminated if the donor P22 phage expressed its Abc (anti-RecBC) function. We hypothesize that in standard (high multiplicity of infection) P22-mediated transduction crosses, recombination is stimulated both by Chi sequences (when present in the transduced fragment) and by the phage-encoded Abc protein which inhibits the host RecBCD exonuclease.
Full text
PDF






Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amundsen S. K., Taylor A. F., Chaudhury A. M., Smith G. R. recD: the gene for an essential third subunit of exonuclease V. Proc Natl Acad Sci U S A. 1986 Aug;83(15):5558–5562. doi: 10.1073/pnas.83.15.5558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barbour S. D., Clark A. J. Biochemical and genetic studies of recombination proficiency in Escherichia coli. I. Enzymatic activity associated with recB+ and recC+ genes. Proc Natl Acad Sci U S A. 1970 Apr;65(4):955–961. doi: 10.1073/pnas.65.4.955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bender J., Kleckner N. Genetic evidence that Tn10 transposes by a nonreplicative mechanism. Cell. 1986 Jun 20;45(6):801–815. doi: 10.1016/0092-8674(86)90555-6. [DOI] [PubMed] [Google Scholar]
- Biek D. P., Cohen S. N. Identification and characterization of recD, a gene affecting plasmid maintenance and recombination in Escherichia coli. J Bacteriol. 1986 Aug;167(2):594–603. doi: 10.1128/jb.167.2.594-603.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burland V., Plunkett G., 3rd, Daniels D. L., Blattner F. R. DNA sequence and analysis of 136 kilobases of the Escherichia coli genome: organizational symmetry around the origin of replication. Genomics. 1993 Jun;16(3):551–561. doi: 10.1006/geno.1993.1230. [DOI] [PubMed] [Google Scholar]
- CLARK A. J., MARGULIES A. D. ISOLATION AND CHARACTERIZATION OF RECOMBINATION-DEFICIENT MUTANTS OF ESCHERICHIA COLI K12. Proc Natl Acad Sci U S A. 1965 Feb;53:451–459. doi: 10.1073/pnas.53.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Botstein D., Watanabe T., Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology. 1972 Dec;50(3):883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- Chaudhury A. M., Smith G. R. A new class of Escherichia coli recBC mutants: implications for the role of RecBC enzyme in homologous recombination. Proc Natl Acad Sci U S A. 1984 Dec;81(24):7850–7854. doi: 10.1073/pnas.81.24.7850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaudhury A. M., Smith G. R. Escherichia coli recBC deletion mutants. J Bacteriol. 1984 Nov;160(2):788–791. doi: 10.1128/jb.160.2.788-791.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciampi M. S., Roth J. R. Polarity effects in the hisG gene of salmonella require a site within the coding sequence. Genetics. 1988 Feb;118(2):193–202. doi: 10.1093/genetics/118.2.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark A. J. Recombination deficient mutants of E. coli and other bacteria. Annu Rev Genet. 1973;7:67–86. doi: 10.1146/annurev.ge.07.120173.000435. [DOI] [PubMed] [Google Scholar]
- Dabert P., Ehrlich S. D., Gruss A. Chi sequence protects against RecBCD degradation of DNA in vivo. Proc Natl Acad Sci U S A. 1992 Dec 15;89(24):12073–12077. doi: 10.1073/pnas.89.24.12073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon D. A., Kowalczykowski S. C. Homologous pairing in vitro stimulated by the recombination hotspot, Chi. Cell. 1991 Jul 26;66(2):361–371. doi: 10.1016/0092-8674(91)90625-9. [DOI] [PubMed] [Google Scholar]
- Dixon D. A., Kowalczykowski S. C. The recombination hotspot chi is a regulatory sequence that acts by attenuating the nuclease activity of the E. coli RecBCD enzyme. Cell. 1993 Apr 9;73(1):87–96. doi: 10.1016/0092-8674(93)90162-j. [DOI] [PubMed] [Google Scholar]
- Dower N. A., Stahl F. W. Chi activity during transduction-associated recombination. Proc Natl Acad Sci U S A. 1981 Nov;78(11):7033–7037. doi: 10.1073/pnas.78.11.7033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dykstra C. C., Prasher D., Kushner S. R. Physical and biochemical analysis of the cloned recB and recC genes of Escherichia coli K-12. J Bacteriol. 1984 Jan;157(1):21–27. doi: 10.1128/jb.157.1.21-27.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebel-Tsipis J., Fox M. S., Botstein D. Generalized transduction by bacteriophage P22 in Salmonella typhimurium. II. Mechanism of integration of transducing DNA. J Mol Biol. 1972 Nov 14;71(2):449–469. doi: 10.1016/0022-2836(72)90362-2. [DOI] [PubMed] [Google Scholar]
- Eichler D. C., Lehman I. R. On the role of ATP in phosphodiester bond hydrolysis catalyzed by the recBC deoxyribonuclease of Escherichia coli. J Biol Chem. 1977 Jan 25;252(2):499–503. [PubMed] [Google Scholar]
- Eisenstark A., Eisenstark R., van Dillewijn J., Rörsch A. Radiation--sensitive and recombinationless mutants of Salmonella typhimurium. Mutat Res. 1969 Nov-Dec;8(3):497–504. doi: 10.1016/0027-5107(69)90066-9. [DOI] [PubMed] [Google Scholar]
- Elliott T., Roth J. R. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol Gen Genet. 1988 Aug;213(2-3):332–338. doi: 10.1007/BF00339599. [DOI] [PubMed] [Google Scholar]
- Emmerson P. T., Howard-Flanders P. Cotransduction with thy of a gene required for genetic recombination in Escherichia coli. J Bacteriol. 1967 May;93(5):1729–1731. doi: 10.1128/jb.93.5.1729-1731.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fenton A. C., Poteete A. R. Genetic analysis of the erf region of the bacteriophage P22 chromosome. Virology. 1984 Apr 15;134(1):148–160. doi: 10.1016/0042-6822(84)90280-0. [DOI] [PubMed] [Google Scholar]
- Goldmark P. J., Linn S. An endonuclease activity from Escherichia coli absent from certain rec- strains. Proc Natl Acad Sci U S A. 1970 Sep;67(1):434–441. doi: 10.1073/pnas.67.1.434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldmark P. J., Linn S. Purification and properties of the recBC DNase of Escherichia coli K-12. J Biol Chem. 1972 Mar 25;247(6):1849–1860. [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Conditionally transposition-defective derivative of Mu d1(Amp Lac). J Bacteriol. 1984 Jul;159(1):130–137. doi: 10.1128/jb.159.1.130-137.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Directed formation of deletions and duplications using Mud(Ap, lac). Genetics. 1985 Feb;109(2):263–282. doi: 10.1093/genetics/109.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner S. R. Differential thermolability of exonuclease and endonuclease activities of the recBC nuclease isolated from thermosensitive recB and recC mutants. J Bacteriol. 1974 Dec;120(3):1219–1222. doi: 10.1128/jb.120.3.1219-1222.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushner S. R. In vivo studies of temperature-sensitive recB and recC mutants. J Bacteriol. 1974 Dec;120(3):1213–1218. doi: 10.1128/jb.120.3.1213-1218.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam S. T., Stahl M. M., McMilin K. D., Stahl F. W. Rec-mediated recombinational hot spot activity in bacteriophage lambda. II. A mutation which causes hot spot activity. Genetics. 1974 Jul;77(3):425–433. doi: 10.1093/genetics/77.3.425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lieberman R. P., Oishi M. The recBC deoxyribonuclease of Escherichia coli: isolation and characterization of the subunit proteins and reconstitution of the enzyme. Proc Natl Acad Sci U S A. 1974 Dec;71(12):4816–4820. doi: 10.1073/pnas.71.12.4816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lovett S. T., Luisi-DeLuca C., Kolodner R. D. The genetic dependence of recombination in recD mutants of Escherichia coli. Genetics. 1988 Sep;120(1):37–45. doi: 10.1093/genetics/120.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahan M. J., Roth J. R. recB and recC genes of Salmonella typhimurium. J Bacteriol. 1989 Jan;171(1):612–615. doi: 10.1128/jb.171.1.612-615.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKittrick N. H., Smith G. R. Activation of Chi recombinational hotspots by RecBCD-like enzymes from enteric bacteria. J Mol Biol. 1989 Dec 5;210(3):485–495. doi: 10.1016/0022-2836(89)90125-3. [DOI] [PubMed] [Google Scholar]
- Murphy K. C., Fenton A. C., Poteete A. R. Sequence of the bacteriophage P22 anti-recBCD (abc) genes and properties of P22 abc region deletion mutants. Virology. 1987 Oct;160(2):456–464. doi: 10.1016/0042-6822(87)90017-1. [DOI] [PubMed] [Google Scholar]
- Oishi M. An ATP-dependent deoxyribonuclease from Escherichia coli with a possible role in genetic recombination. Proc Natl Acad Sci U S A. 1969 Dec;64(4):1292–1299. doi: 10.1073/pnas.64.4.1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver D. B., Goldberg E. B. Protection of parental T4 DNA from a restriction exonuclease by the product of gene 2. J Mol Biol. 1977 Nov;116(4):877–881. doi: 10.1016/0022-2836(77)90276-5. [DOI] [PubMed] [Google Scholar]
- Palas K. M., Kushner S. R. Biochemical and physical characterization of exonuclease V from Escherichia coli. Comparison of the catalytic activities of the RecBC and RecBCD enzymes. J Biol Chem. 1990 Feb 25;265(6):3447–3454. [PubMed] [Google Scholar]
- Poteete A. R., Fenton A. C., Murphy K. C. Modulation of Escherichia coli RecBCD activity by the bacteriophage lambda Gam and P22 Abc functions. J Bacteriol. 1988 May;170(5):2012–2021. doi: 10.1128/jb.170.5.2012-2021.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinken R., Thomas B., Wackernagel W. Evidence that recBC-dependent degradation of duplex DNA in Escherichia coli recD mutants involves DNA unwinding. J Bacteriol. 1992 Aug;174(16):5424–5429. doi: 10.1128/jb.174.16.5424-5429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roof D. M., Roth J. R. Ethanolamine utilization in Salmonella typhimurium. J Bacteriol. 1988 Sep;170(9):3855–3863. doi: 10.1128/jb.170.9.3855-3863.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg S. M., Hastings P. J. The split-end model for homologous recombination at double-strand breaks and at Chi. Biochimie. 1991 Apr;73(4):385–397. doi: 10.1016/0300-9084(91)90105-a. [DOI] [PubMed] [Google Scholar]
- Russell C. B., Thaler D. S., Dahlquist F. W. Chromosomal transformation of Escherichia coli recD strains with linearized plasmids. J Bacteriol. 1989 May;171(5):2609–2613. doi: 10.1128/jb.171.5.2609-2613.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmieger H. A method for detection of phage mutants with altered transducing ability. Mol Gen Genet. 1971;110(4):378–381. doi: 10.1007/BF00438281. [DOI] [PubMed] [Google Scholar]
- Shyamala V., Schneider E., Ames G. F. Tandem chromosomal duplications: role of REP sequences in the recombination event at the join-point. EMBO J. 1990 Mar;9(3):939–946. doi: 10.1002/j.1460-2075.1990.tb08192.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith G. R., Amundsen S. K., Chaudhury A. M., Cheng K. C., Ponticelli A. S., Roberts C. M., Schultz D. W., Taylor A. F. Roles of RecBC enzyme and chi sites in homologous recombination. Cold Spring Harb Symp Quant Biol. 1984;49:485–495. doi: 10.1101/sqb.1984.049.01.055. [DOI] [PubMed] [Google Scholar]
- Stahl F. W., Thomason L. C., Siddiqi I., Stahl M. M. Further tests of a recombination model in which chi removes the RecD subunit from the RecBCD enzyme of Escherichia coli. Genetics. 1990 Nov;126(3):519–533. doi: 10.1093/genetics/126.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl M. M., Kobayashi I., Stahl F. W., Huntington S. K. Activation of Chi, a recombinator, by the action of an endonuclease at a distant site. Proc Natl Acad Sci U S A. 1983 Apr;80(8):2310–2313. doi: 10.1073/pnas.80.8.2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor A. F., Smith G. R. Substrate specificity of the DNA unwinding activity of the RecBC enzyme of Escherichia coli. J Mol Biol. 1985 Sep 20;185(2):431–443. doi: 10.1016/0022-2836(85)90414-0. [DOI] [PubMed] [Google Scholar]
- Thaler D. S., Sampson E., Siddiqi I., Rosenberg S. M., Thomason L. C., Stahl F. W., Stahl M. M. Recombination of bacteriophage lambda in recD mutants of Escherichia coli. Genome. 1989;31(1):53–67. doi: 10.1139/g89-013. [DOI] [PubMed] [Google Scholar]
- Tomizawa J., Ogawa H. Structural genes of ATP-dependent deoxyribonuclease of Escherichia coli. Nat New Biol. 1972 Sep 6;239(88):14–16. doi: 10.1038/newbio239014a0. [DOI] [PubMed] [Google Scholar]
- Triman K. L., Chattoraj D. K., Smith G. R. Identity of a Chi site of Escherichia coli and Chi recombinational hotspots of bacteriophage lambda. J Mol Biol. 1982 Jan 15;154(2):393–399. doi: 10.1016/0022-2836(82)90072-9. [DOI] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Willetts N. S., Mount D. W. Genetic analysis of recombination-deficient mutants of Escherichia coli K-12 carrying rec mutations cotransducible with thyA. J Bacteriol. 1969 Nov;100(2):923–934. doi: 10.1128/jb.100.2.923-934.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]