Abstract
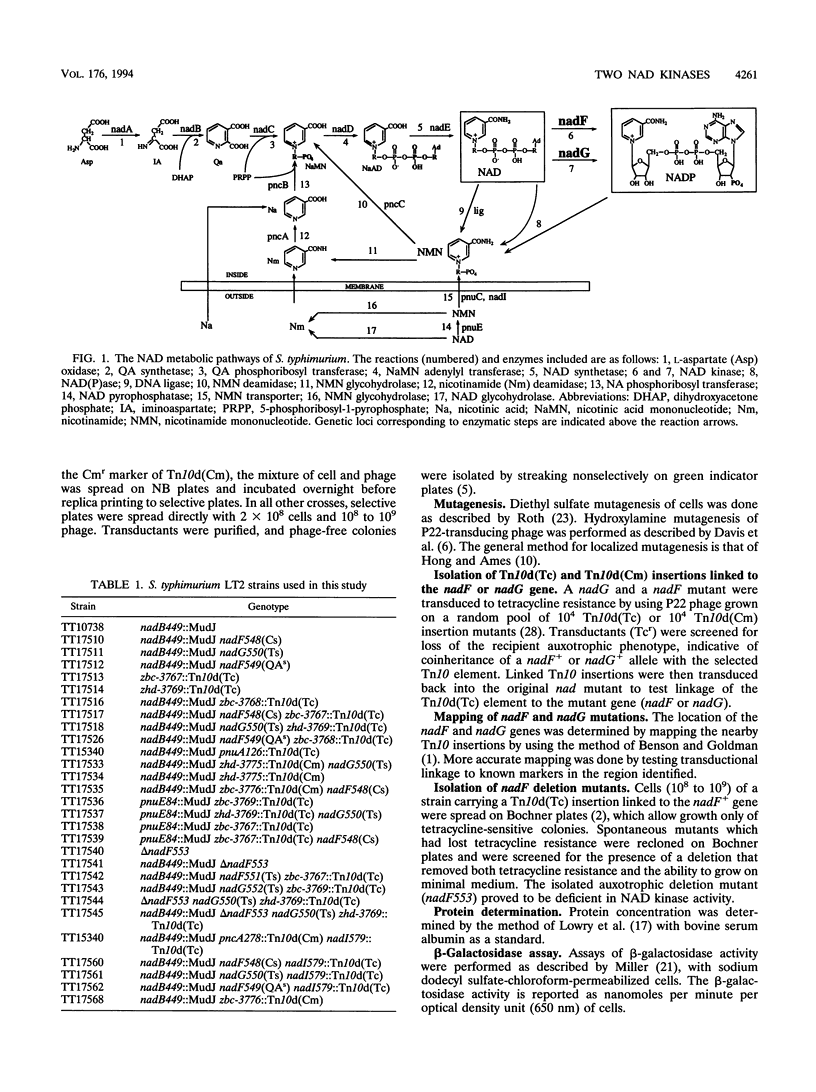
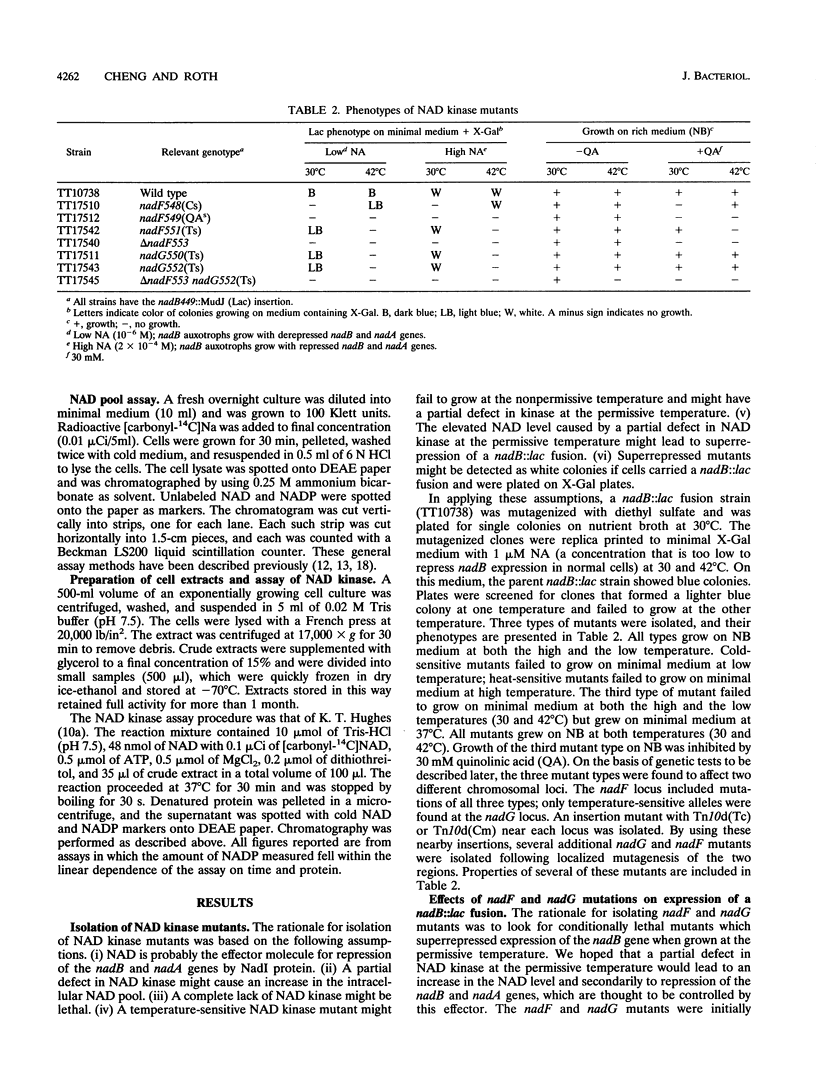
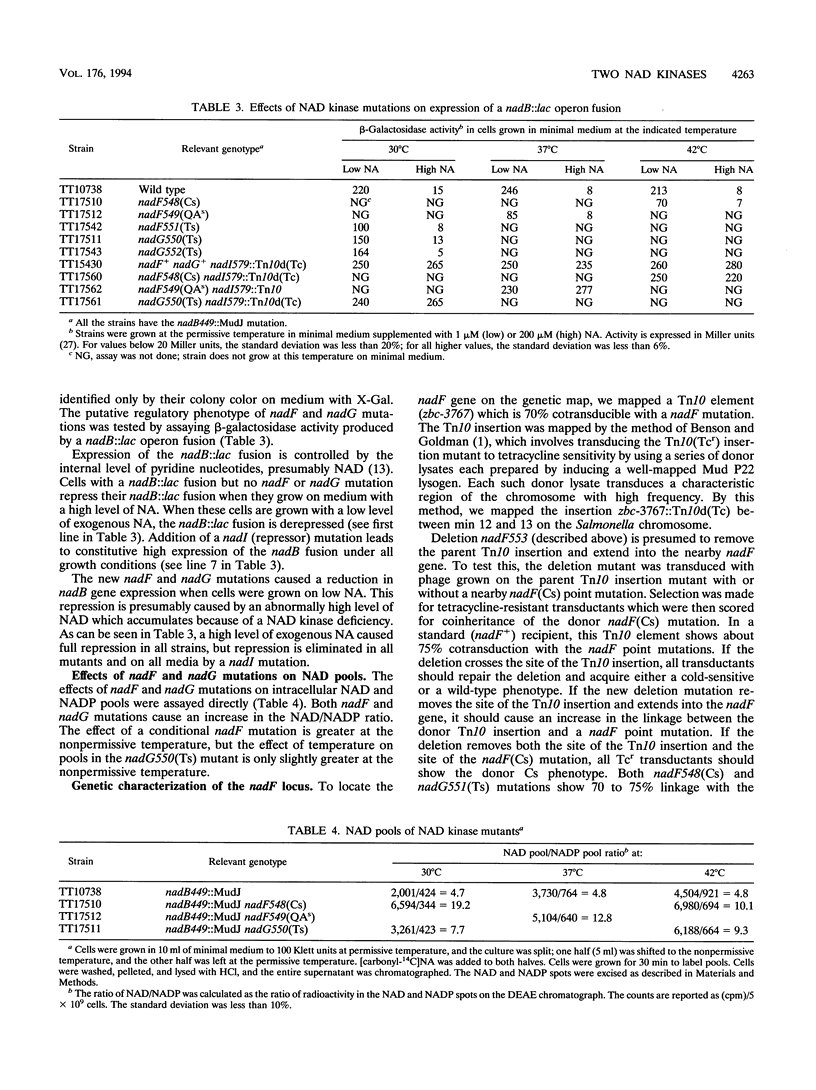
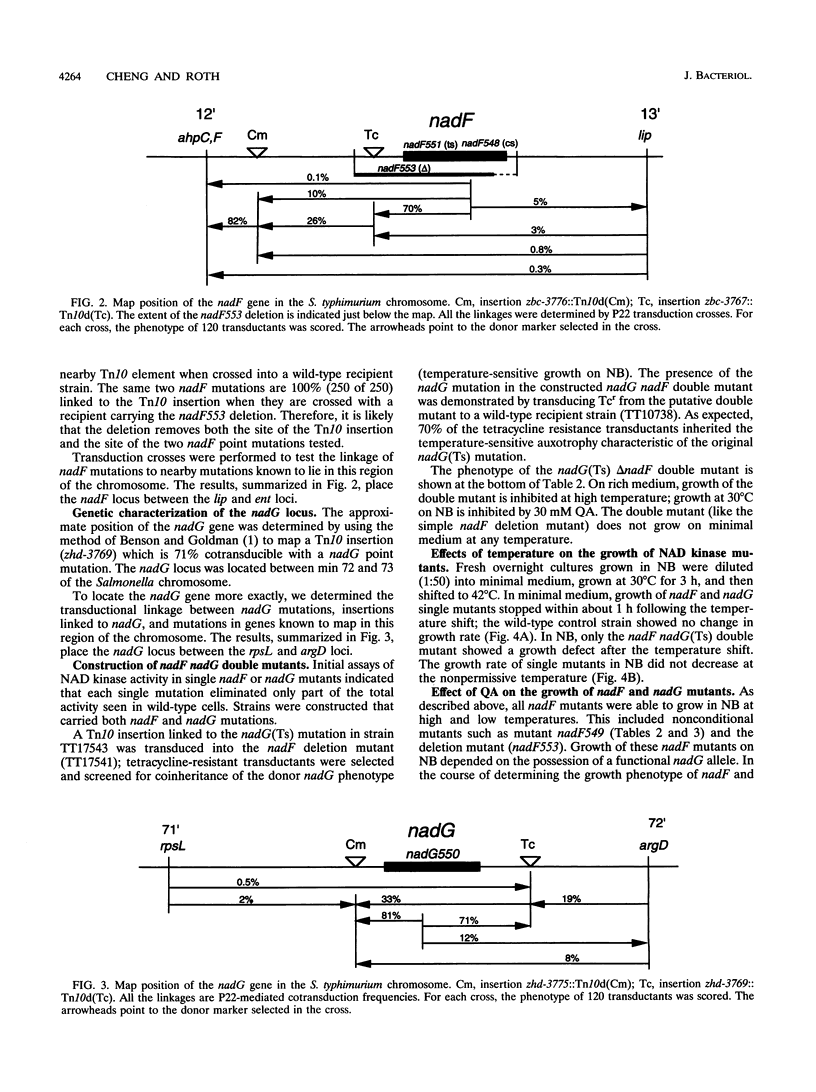
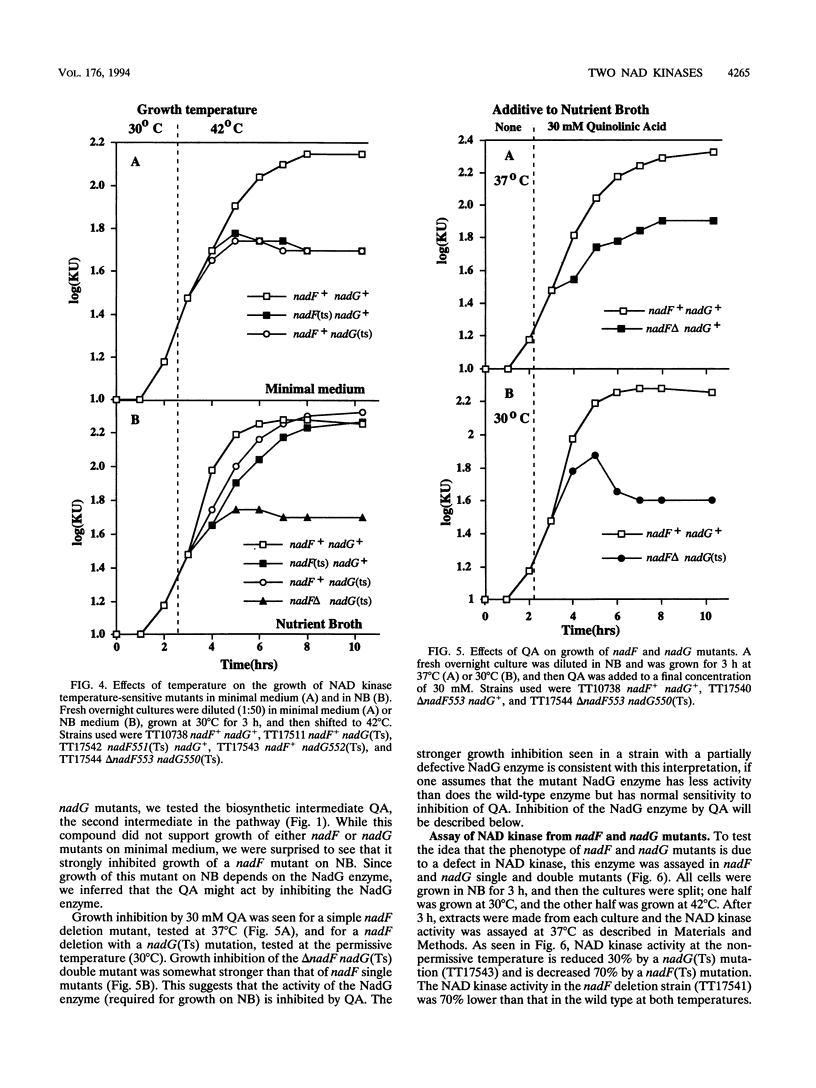
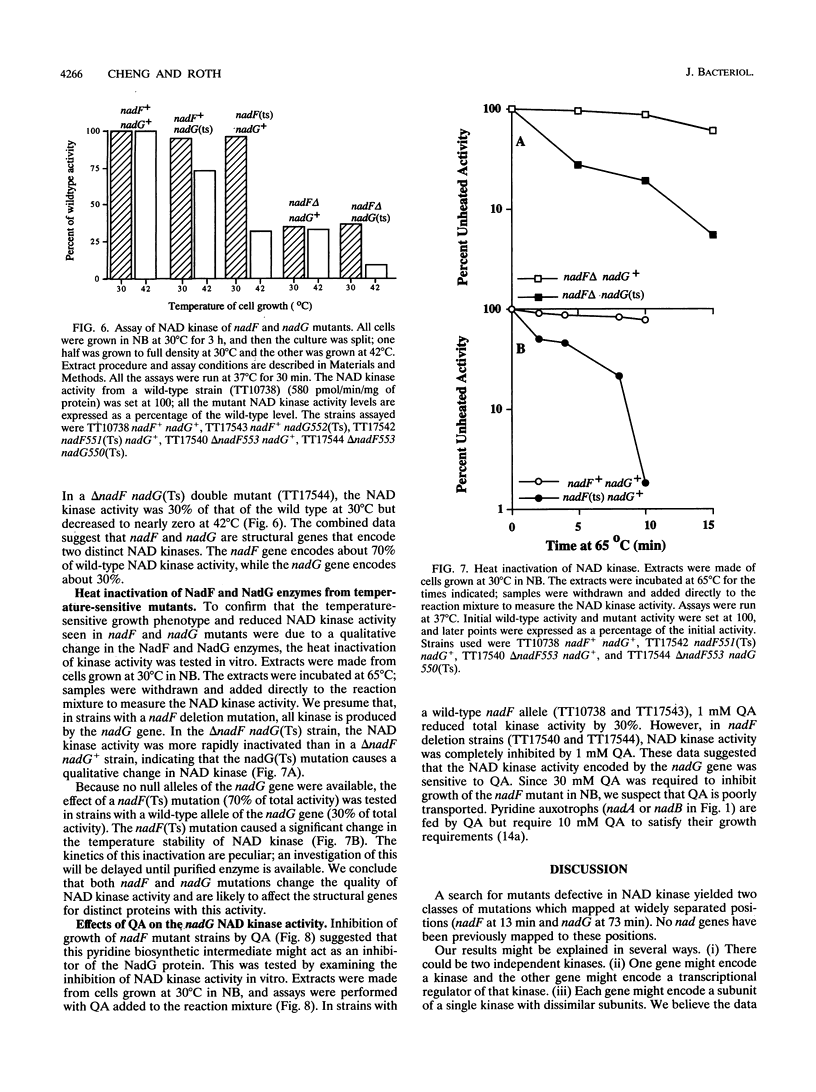
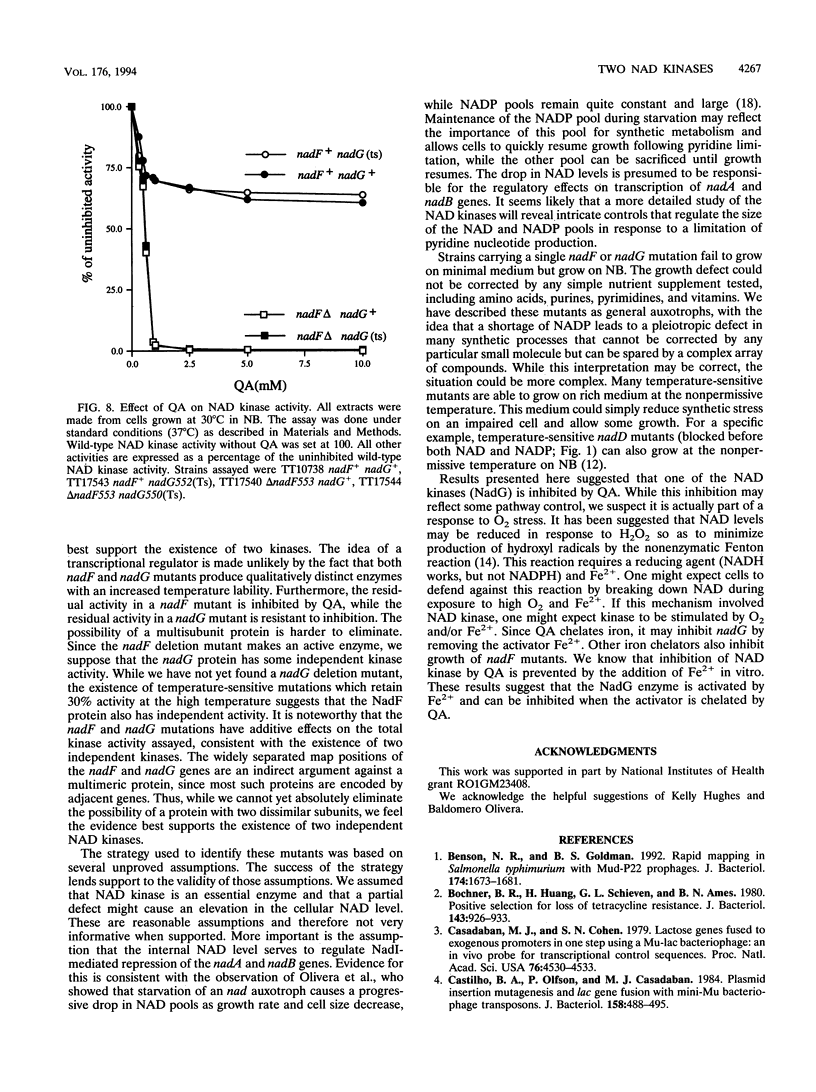
The electron-carrying cofactor NADP is formed by phosphorylation of NAD. A strategy for the isolation of NAD kinase mutants revealed two classes of temperature-sensitive mutations, nadF and nadG, mapping at min 13 and 72 of the Salmonella chromosome. Both mutant types grew on nutrient broth at both 30 and 42 degrees C but on minimal medium showed a temperature-sensitive growth defect which was not corrected by any of the single nutritional supplements tested. A nadF deletion mutant grew on nutrient broth but not on minimal medium. A double mutant with the nadF deletion and a nadG(Ts) mutation showed temperature-sensitive growth on all media. We propose that Salmonella typhimurium has two NAD kinases, one encoded by the nadF and one by the nadG gene. This is supported by the fact that temperature-sensitive mutants of both genes produce kinase activity with altered heat stability. Results suggest that either one of two NAD kinases is sufficient for growth on rich medium, but that both are needed for growth on minimal media. Enzyme assays show that the nadF gene is responsible for about 70% of total NAD kinase activity, and that the nadG gene dictates the remaining 30%. While testing nutritional phenotypes of nadF and nadG mutants, we found that the biosynthetic intermediate, quinolinic acid (QA) inhibited growth of nadF mutants on nutrient broth. This suggested that the NadG enzyme might be inhibited by QA. Enzyme assays demonstrated that QA inhibits the NadG but not the NadF enzyme. This suggests the existence of a regulatory mechanism which controls NADP levels.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Benson N. R., Goldman B. S. Rapid mapping in Salmonella typhimurium with Mud-P22 prophages. J Bacteriol. 1992 Mar;174(5):1673–1681. doi: 10.1128/jb.174.5.1673-1681.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bochner B. R., Huang H. C., Schieven G. L., Ames B. N. Positive selection for loss of tetracycline resistance. J Bacteriol. 1980 Aug;143(2):926–933. doi: 10.1128/jb.143.2.926-933.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casadaban M. J., Cohen S. N. Lactose genes fused to exogenous promoters in one step using a Mu-lac bacteriophage: in vivo probe for transcriptional control sequences. Proc Natl Acad Sci U S A. 1979 Sep;76(9):4530–4533. doi: 10.1073/pnas.76.9.4530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castilho B. A., Olfson P., Casadaban M. J. Plasmid insertion mutagenesis and lac gene fusion with mini-mu bacteriophage transposons. J Bacteriol. 1984 May;158(2):488–495. doi: 10.1128/jb.158.2.488-495.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan R. K., Botstein D., Watanabe T., Ogata Y. Specialized transduction of tetracycline resistance by phage P22 in Salmonella typhimurium. II. Properties of a high-frequency-transducing lysate. Virology. 1972 Dec;50(3):883–898. doi: 10.1016/0042-6822(72)90442-4. [DOI] [PubMed] [Google Scholar]
- Elliott T., Roth J. R. Characterization of Tn10d-Cam: a transposition-defective Tn10 specifying chloramphenicol resistance. Mol Gen Genet. 1988 Aug;213(2-3):332–338. doi: 10.1007/BF00339599. [DOI] [PubMed] [Google Scholar]
- Foster J. W., Moat A. G. Mapping and characterization of the nad genes in Salmonella typhimurium LT-2. J Bacteriol. 1978 Feb;133(2):775–779. doi: 10.1128/jb.133.2.775-779.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Foster J. W., Moat A. G. Nicotinamide adenine dinucleotide biosynthesis and pyridine nucleotide cycle metabolism in microbial systems. Microbiol Rev. 1980 Mar;44(1):83–105. doi: 10.1128/mr.44.1.83-105.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong J. S., Ames B. N. Localized mutagenesis of any specific small region of the bacterial chromosome. Proc Natl Acad Sci U S A. 1971 Dec;68(12):3158–3162. doi: 10.1073/pnas.68.12.3158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Ladika D., Roth J. R., Olivera B. M. An indispensable gene for NAD biosynthesis in Salmonella typhimurium. J Bacteriol. 1983 Jul;155(1):213–221. doi: 10.1128/jb.155.1.213-221.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Olivera B. M., Roth J. R. Structural gene for NAD synthetase in Salmonella typhimurium. J Bacteriol. 1988 May;170(5):2113–2120. doi: 10.1128/jb.170.5.2113-2120.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes K. T., Roth J. R. Conditionally transposition-defective derivative of Mu d1(Amp Lac). J Bacteriol. 1984 Jul;159(1):130–137. doi: 10.1128/jb.159.1.130-137.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imlay J. A., Chin S. M., Linn S. Toxic DNA damage by hydrogen peroxide through the Fenton reaction in vivo and in vitro. Science. 1988 Apr 29;240(4852):640–642. doi: 10.1126/science.2834821. [DOI] [PubMed] [Google Scholar]
- LOWRY O. H., ROSEBROUGH N. J., FARR A. L., RANDALL R. J. Protein measurement with the Folin phenol reagent. J Biol Chem. 1951 Nov;193(1):265–275. [PubMed] [Google Scholar]
- Lehman I. R. DNA ligase: structure, mechanism, and function. Science. 1974 Nov 29;186(4166):790–797. doi: 10.1126/science.186.4166.790. [DOI] [PubMed] [Google Scholar]
- Lundquist R., Olivera B. M. Pyridine nucleotide metabolism in Escherichia coli. II. Niacin starvation. J Biol Chem. 1973 Jul 25;248(14):5137–5143. [PubMed] [Google Scholar]
- McGuinness E. T., Butler J. R. NAD+ kinase--a review. Int J Biochem. 1985;17(1):1–11. doi: 10.1016/0020-711x(85)90079-5. [DOI] [PubMed] [Google Scholar]
- Olivera B. M., Lehman I. R. Diphosphopyridine nucleotide: a cofactor for the polynucleotide-joining enzyme from Escherichia coli. Proc Natl Acad Sci U S A. 1967 Jun;57(6):1700–1704. doi: 10.1073/pnas.57.6.1700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritz G. J., Chandler J. L. Recognition of a gene involved in the regulation of nicotinamide adenine dinucleotide biosynthesis. J Bacteriol. 1973 Apr;114(1):128–136. doi: 10.1128/jb.114.1.128-136.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VOGEL H. J., BONNER D. M. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956 Jan;218(1):97–106. [PubMed] [Google Scholar]
- WANG T. P., KAPLAN N. O. Kinases for the synthesis of coenzyme A and triphosphopyridine nucleotide. J Biol Chem. 1954 Jan;206(1):311–325. [PubMed] [Google Scholar]
- Way J. C., Davis M. A., Morisato D., Roberts D. E., Kleckner N. New Tn10 derivatives for transposon mutagenesis and for construction of lacZ operon fusions by transposition. Gene. 1984 Dec;32(3):369–379. doi: 10.1016/0378-1119(84)90012-x. [DOI] [PubMed] [Google Scholar]
- Zerez C. R., Moul D. E., Andreoli A. J. NAD kinase from Bacillus licheniformis: inhibition by NADP and other properties. Arch Microbiol. 1986 May;144(4):313–316. doi: 10.1007/BF00409878. [DOI] [PubMed] [Google Scholar]
- Zerez C. R., Moul D. E., Gomez E. G., Lopez V. M., Andreoli A. J. Negative modulation of Escherichia coli NAD kinase by NADPH and NADH. J Bacteriol. 1987 Jan;169(1):184–188. doi: 10.1128/jb.169.1.184-188.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]


