Abstract
Baculovirus occlusion-derived virus (ODV) derives its envelope from an intranuclear membrane source. N-terminal amino acid sequences of the Autographa californica nuclear polyhedrosis virus (AcMNPV) envelope proteins, ODV-E66 and ODV-E25 (23 and 24 amino acids, respectively) are highly hydrophobic. Recombinant viruses that express the two N-terminal amino acid sequences fused to green fluorescent protein (23GFP or 24GFP) provided visual markers to follow protein transport and localization within the nucleus during infection. Autoflourescence was first detected along the cytoplasmic periphery of the nucleus and subsequently localized as foci to discrete locations within the nucleus. Immunoelectron microscopy confirmed that these foci predominantly contained intranuclear microvesicles and the reporter fusion proteins were also detected in cytoplasmic membranes near the nucleus, and the outer and inner nuclear membrane. Therefore, these defined hydrophobic domains are sufficient to direct native and fusion proteins to induced membrane microvesicles within a baculovirus-infected cell nucleus and the viral envelope. In addition, these data suggest that movement of these proteins into the nuclear envelope may initiate through cytoplasmic membranes, such as endoplasmic reticulum, and that transport into the nucleus may be mediated through the outer and inner nuclear membrane.
The Autographa californica nuclear polyhedrosis virus (AcMNPV) infection of insect cells produces two enveloped progeny viruses: budded virus and occlusion-derived virus (ODV). The budded virus obtains its envelope from the cell surface when the nucleocapsid buds through the plasma membrane as it exits the cell. This strategy is similar to that observed for other viruses that bud from the cell surface (1). The molecular mechanism utilized by ODV to obtain an envelope within the nucleus is unlike other viral nuclear maturation strategies. ODV obtains its envelope from viral induced membranes within the nucleoplasm. The mature ODV is then occluded within a proteinaceous crystal matrix and upon lysis of the infected cells, occlusions are released into the environment to be consumed by another insect (2, 3).
Infection by AcMNPV and other baculoviruses induce an extensive elaboration and proliferation of intranuclear membranes that appear as microvesicles and unit membrane structures within the nucleoplasm (4, 5). Previous results using ODV envelope proteins as markers to study trafficking and assembly of intranuclear microvesicles and the ODV envelope, suggest that movement of these proteins could be mediated through cytoplasmic membranes and the nuclear envelope (6). This model predicts that ODV envelope proteins are incorporated into the endoplasmic reticulum (ER), and transported to the outer and inner nuclear membrane (ONM and INM). This model does not rule out nuclear pore import and subsequent sorting to the INM or induced microvesicles (6). Our data suggest that ODV envelope proteins become incorporated, at least transiently with the INM. Based on these observations, one might expect ODV envelope proteins would contain targeting/retention signals similar to other INM proteins. The molecular signals necessary for transport or retention of proteins into the INM are largely uncharacterized; however, for certain proteins a hydrophobic domain has been identified to play a role. Transmembrane domains from lamin B receptor (LBR), glycoprotein (gp) 210, and herpes simplex virus glycoprotein B can direct proteins to the nuclear envelope (7–9). It is unknown why these domains are sufficient to direct proteins to the nuclear envelope while other, apparently similar transmembrane domains do not function as such.
The availability of several viral envelope protein markers provides the opportunity to use baculovirus infection to study protein movement into the nuclear envelope and into membranes that are induced to proliferate and subsequently assemble as viral envelopes within the nucleus. Such a system offers several advantages for studying these processes including the following: (i) virus infection amplifies protein trafficking and movement into nuclear membranes thus making the process easier to detect and follow; (ii) the timing of expression for the envelope protein or reporter constructs can be controlled using viral promoters of immediate early, late, and very late classes; (iii) insect cells are similar in many respects to mammalian cells, thus pathways and mechanisms elucidated utilizing this model give insights to similar pathways of mammalian cells; and (iv) the use of marker proteins and baculovirus provides an opportunity to trace the movement of membranes and their associated proteins into the nucleus and nucleoplasm. In this study, we use a combination of unique viral envelope protein markers, and fusion proteins thereof, to investigate protein transport and localization to the nuclear envelope, membranes within the nucleoplasm and the viral envelope.
MATERIALS AND METHODS
ODV-E66 Constructs.
The plasmid construct Δ2–23β-galactosidase (β-gal) (Fig. 1A, row 2) was generated using the transformer site-directed mutagenesis kit (CLONTECH). Amino acids 2–22 were deleted using the deletion oligonucleotide 5′-GCAACATTCGACATGAGCAATAATAAAAATGATGCC-3′ and the selection oligonucleotide 5′-CTCTAGAGGAACCCCGGGAACCGAGCTCG-3′. After the introduction of deletion, the KpnI–KpnI fragment (28.1–28.2 map units) was replaced with a 3.1-kb KpnI fragment containing the LacZ gene from a previously generated clone 125β-gal (T.H., unpublished data).
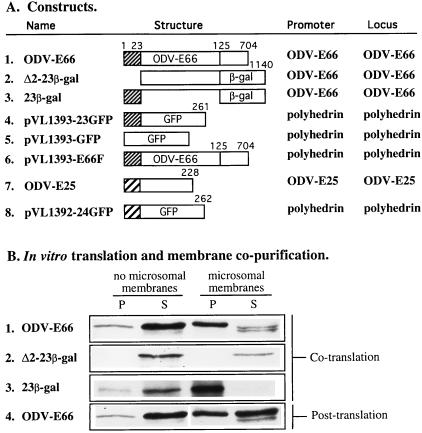
Figure 1.
Genetic constructs, in vitro translation, and membrane copurification. (A) Genetic constructs. Numbers indicate positions of amino acids based upon ODV-E66. (B) In vitro translation and membrane copurification. P, microsomal membrane pellet fraction; S, supernatant fraction.
To obtain the construct 23β-gal (Fig. 1A, row 3), nucleotide sequence encoding amino acids 24–25 were mutated to a KpnI site using the selection oligonucleotide 5′-CTCTAGAGGAACCCCGGGAACCGAGCTCG-3′ and the site-mutation oligonucleotide 5′-CCTATCAAATAGC GGTACCAAAAATGATGCCAAT-3′ (KpnI site is underlined). The region from the new KpnI site at amino acids 24 and 25 to the KpnI site at 28.2 map units was replaced with the 3.1-kb KpnI fragment containing the LacZ gene from 125β-gal.
The construct 23-green fluorescent protein (GFP) (Fig. 1A, row 4) was obtained by amplifying the N-terminal 23 amino acids of ODV-E66 by PCR using the following oligonucleotides: 5′-TTTTTTAAGCTTATGTCTATCGTATTG-3′ (HindIII) and 5′-TTTTTTGGATCCTTGCTATTTGATAGGTA-3′ (BamHI). The PCR product was digested and cloned into pUC19. The N-terminal 23 amino acids of ODV-E66 were fused in frame with GFP (CLONTECH; pUC19–23GFP). This construct was then cloned into the baculovirus transfer vector, pVL1393 (ref. 10; pVL 1393–23GFP).
To express ODV-E66 under the control of the polyhedrin promoter (Fig. 1A, row 6), ODV-E66 was amplified by PCR using the following oligonucleotides: 5′-TTTTTGGATCCACC ATGTCTATCGTATTG-3′ (BamHI) and 5′-TTTTTCTGCAGTTACTTGTCGTCGTCGTCTTTGTAGTCCACAATTTCAAAAAT-3′ (PstI) and cloned into pVL1393 (pVL1393-E66F) .
ODV-E25 Constructs.
To generate 24GFP (Fig. 1A, row 8) the N-terminal 24 amino acids of ODV-E25 were amplified by PCR using the following oligonucleotides: 5′-TTTTTTCTGCAGATGTGGGGAATCGTG-3′ and 5′-TTTTTTGGATCCTTGAAATTTAATGCATT-3′. The PCR product digested with PstI and BamHI and the BamHI/EcoRI fragment encoding GFP from pGFP vector were cloned into PstI/EcoRI sites of pVL1392.
All cloning constructs were verified by DNA sequencing.
Cells and Construction of Recombinant Viruses.
Recombinant viruses Δ2–23β-gal and 23β-gal were constructed using the corresponding transfer plasmid and AcMNPV viral DNA (11). pVL1393–23GFP, pVL1393-E66F, and pVL1392–24GFP were constructed using the corresponding transfer plasmid and Bsu36I-digested BakPak6 viral DNA (CLONTECH). The locus of recombination was verified by Southern blot analyses and appropriate probes. Protein expression was confirmed by Western blot analyses of infected cell extracts using appropriate antibody: β-gal recombinant viruses (mouse anti-β-gal polyclonal antibody at 1:1250; Sigma); 23GFP recombinant viruses (rabbit anti-GFP polyclonal antibody at 1:1500; CLONTECH). Southern and Western blot analyses were done as described by Sambrook et al. (12). Spodoptera frugiperda (Sf9) cells were infected at a multiplicity of infection of 20.
In Vitro Transcription, Translation, and Membrane Copurification Assay.
Genes encoding ODV-E66, Δ2–23β-gal, and 23β-gal were cloned into pBS− vector. Plasmid DNA was linearized and transcribed (5). Cotranslational membrane copurification assay was done as described by Kabcenell and Atkinson (13). Briefly, after translation membranes were diluted with a high salt buffer (20 mM Tris·HCl/500 mM KCl/2 mM CaCl2/5 mM MgCl2, pH 7.4) and layered over a sucrose cushion (0.5 M) made in the same buffer. Posttranslational membrane copurification was performed after translation for 1 hr at 30°C in the absence of microsomal membranes, 2 mM cycloheximide was added to stop translation, and the reaction mixture was chilled on ice before addition of 1.8 μl of microsomal membranes. After 1-hr incubation at 4°C, the mixture was subject to the membrane copurification assay briefly described (13).
RESULTS
We observed that all the known ODV envelope proteins, ODV-E66, ODV-E25, ODV-E56, and ODV-E18 contain at least one putative hydrophobic domain (5, 6, 14, 15). The hydrophobic domain of ODV-E66 and ODV-E25 is located at the N terminus. N-terminal amino acid sequencing revealed that these proteins are uncleaved in the ODV envelope (see Fig. 4). ODV-E66 and fusion proteins containing the ODV-E66 N-terminal hydrophobic domain were tested for their ability to stably integrate into microsomal membranes. ODV-E66 and 23β-gal proteins were able to incorporate into microsomal membranes (Fig. 1B, rows 1 and 3), whereas Δ2–23β-gal remained in the soluble fraction (Fig. 1B, row 2). Additionally, we determined that ODV-E66 was capable of posttranslational insertion into microsomal membranes, however at a lower efficiency (Fig. 1B, row 4). To determine if ODV-E66 was transported into the lumen of the microsomal membranes, protease digestion experiments were performed and these experiments showed that after insertion, ODV-E66 was not protected from protease activity. These data are consistent with glycosylation studies that indicate that in the mature virus ODV-E66 is not N-glycosylated (data not shown).
Figure 4.
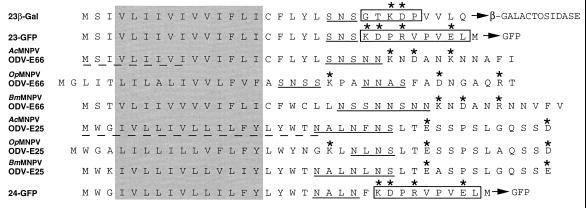
Comparison of N-terminal domains of ODV-E66, ODV-E25, and fusion constructs. Blocked and shaded amino acids indicate hydrophobic domain; underlined (dashed) amino acids were directly N-terminal sequenced; underlined (solid) amino acids indicate N-, S-rich region, boxed amino acids indicate amino acids added by cloning strategy; asterisk indicates charged amino acids. OpMNPV, Orgyia pseudotsugata nuclear polyhedrosis virus; BmMNPV, Bombyx mori nuclear polyhedrosis virus.
One of the unique features of utilizing AcMNPV to express foreign genes is that the temporal pattern and the level of gene expression can be controlled during the infection process by using different viral gene promoters. Two recombinant viruses were generated that contained the N-terminal 23 amino acids of ODV-E66 fused in frame with GFP (23GFP) or β-gal (23β-gal; Fig. 1A, rows 3 and 4). By using either the native or polyhedrin gene promoters, these fusion proteins are expressed at different levels and at different times during infection. The 23β-gal fusion replaced ODV-E66 in its native gene locus and was expressed with the ODV-E66 promoter. Thus expression of 23β-gal should parallel that of ODV-E66 and the cellular localization and abundance of this protein should also be similar. 23GFP was expressed under the control of the polyhedrin promoter. Expression of this gene is at a much higher level, and continues later in the infection process when compared with ODV-E66. Additionally, in this recombinant virus ODV-E66 remains in its native gene locus. Thus, 23GFP functions as a highly expressed, autofluorescent marker where localization can be visually traced.
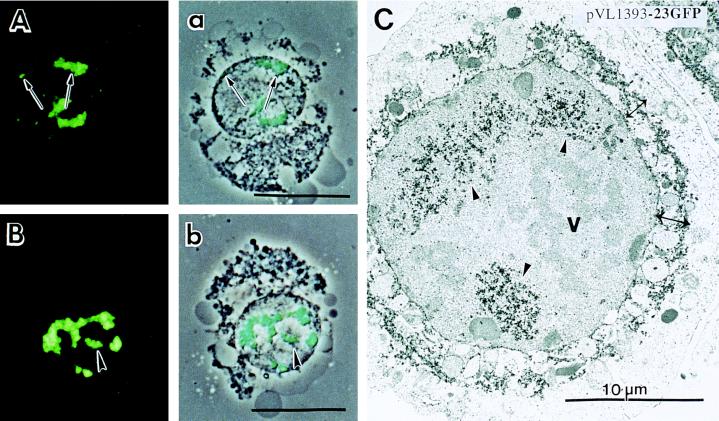
Autofluorescence reveals that 23GFP first accumulates at the periphery of the nucleus at 48 hr postinfection (Fig. 2 A and a, →) and later in infection concentrates at interior regions of the nucleus in discrete foci (72 hr postinfection; Fig. 2 B and b, ➤). Fig. 2C is an immunoelectron micrograph of an infected cell (48 hr postinfection) showing that cytoplasmic membranes condense around the periphery of the nucleus and are labeled using antibody to GFP and gold particles linked to secondary antibody (↔). 23GFP also locates to intranuclear foci of viral-induced membranes (Fig. 2C, ➤). The autofluorescence of GFP allowed us to visually screen large numbers of infected cells and over the course of infection the trafficking and localization pattern of 23GFP was highly reproducible. This pattern included the following. (i) Accumulation at the periphery of the nuclear envelope. In many examples a ring of autofluorescence around the nucleus was very strong as one might predict from the intense labeling demonstrated in Fig. 2C. At the light level we could discriminate autofluorescence in the cytoplasm adjacent to the nuclear envelope, along the interior region of the nuclear envelope and within the nucleus. (ii) 23GFP localized in the nucleus as discrete foci. Because the pattern of fluorescence was never diffuse in the nucleus, and we could always discern discrete regions of accumulation of 23GFP vs. other viral structures, we conclude that the assembly process for the baculovirus envelope within the nucleus is compartmentalized. (iii) When 23GFP was placed in an occlusion positive recombinant virus, the viral occlusions appeared greenish, with a fluorescent tinge showing that the virus containing 23GFP in its envelope was incorporated within occlusions in a normal manner (immunoelectron microscopy data confirmed these results; data not shown). As control, a recombinant virus expressing wild-type GFP in the polyhedrin locus was studied (12, 13). When not linked to the ODV-E66 N-terminal sequence, autofluorescence and immunoelectron microscopy revealed that GFP was uniformly present throughout the cytoplasm and nucleus (data not shown). Thus the discrete patterns of label of 23GFP localization are due to the fused viral envelope N-terminal domain.
Figure 2.
Fluorescence and ImmunoGold (Janssen) localization of pVL1393–23GFP infected cells. (A and B) 23GFP autofluorescence in pVL1393-23GFP-infected Sf9 cells. (a and b) GFP autofluorescence and phase contract double exposure. (A and a) Forty-eight hours postinfection. (B and b) Seventy-two hours postinfection. Exposure time for cells at different time points postinfection was constant and set for exposure of the 72-hr postinfection. (C) ImmunoGold labeling (48 hr postinfection) using antiserum to GFP (1:1500; CLONTECH) and secondary gold conjugate. Fixation and ImmunoGold labeling of infected were done as described in Hong et al. (5). ( ), Intranuclear microvesicles, (↔), cytoplasmic membranes condensed near the nuclear envelope. Protein was detected as in C. C, cytoplasm; n, nucleus.
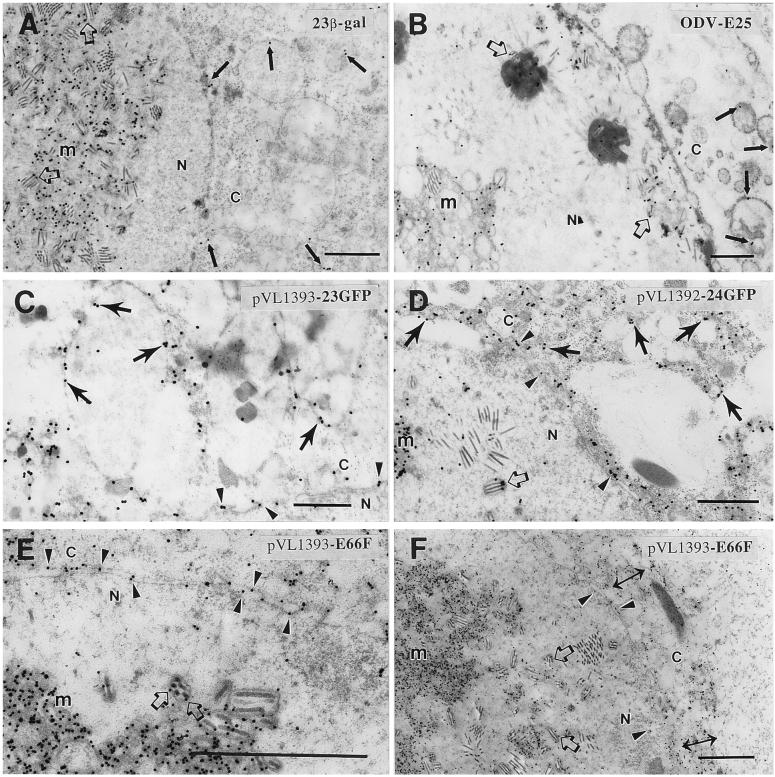
The 23β-gal recombinant virus expressed the fusion protein under the native ODV-E66 promoter. Using antibodies to β-gal, the localization pattern of 23β-gal by immunoelectron microscopy was similar to the wild-type ODV-E66 that was described previously (5). This included significant levels of label within the viral induced intranuclear microvesicles (Fig. 3 A, m), ODV envelope (Fig. 3A, ➯) and slight but discernible label with the nuclear envelope and associated with intracellular, cytoplasmic membranes (Fig. 3A, →). This labeling pattern is very similar to ODV-E25, another ODV envelope protein that contains an N-terminal hydrophobic domain (15). Fig. 3B shows that ODV-E25 antibody also labels intranuclear microvesicles and the ODV envelope (Fig. 3B, m and ➯) with limited but discernible labeling of the nuclear envelope and intracellular cytoplasmic membranes (Fig. 3B, →). When compared with the labeling pattern of wild-type or fusion proteins expressed under the ODV-E66 promoter, fusion proteins more highly expressed under the control of the polyhedrin promoter still label the viral induced microvesicles and ODV envelope, but significant amounts of label are now visualized associated with intracellular cytoplasmic membranes located near the periphery of the nucleus (Fig. 3 C–F). Increased amounts of label are also visualized in the microvesicles and ODV envelope (Fig. 3 D–F, m and ➯). However, label is enhanced in the nuclear envelope, both outer and INM (Fig. 3 D–F, ➤) and in cytoplasmic membranes near the nucleus (Fig. 3C, D, F ↔ and →).
Figure 3.
ImmunoGold localization of ODV-E25, 23β-gal, 23GFP, 24GFP, and ODV-E66. All data are presented from Sf9 infected cells at 48 hr postinfection and secondary antibody linked to gold. (A) 23β-gal recombinant virus (anti-β-gal); (B) AcMNPV (E2 strain; anti-ODV-E25 1:1000). (C) 23GFP recombinant virus (anti-GFP). (D) 24GFP recombinant virus (anti-GFP). (E) pVL-E66 recombinant virus (anti-ODV-E66, ref. 5). n, nucleus; C, cytoplasm; m, microvesicles; ➯, labeling of ODV envelope; ➤, labeling of the nuclear envelope; →, labeling of cytoplasmic membranes; ↔, condensation of cytoplasmic membranes to the periphery of the nucleus. (Bar = 1 μm.)
We compared the N-terminal amino acid sequence of ODV-E66 and ODV-E25 with the sequence of these proteins from other baculoviruses and the fusion constructs generated for this study (Fig. 4). These sequences contain a hydrophobic domain composed primarily of isoleucine, leucine, and valine followed by a asparagine-serine rich region and finally a region containing charged amino acids.
DISCUSSION
In AcMNPV Infection, the Nucleus Becomes Significantly Enlarged.
Transmission electron microscopy reveals that although apparently remaining intact, regions of the nuclear envelope and INM become irregular (18). As infection progresses, foci of microvesicles appear and localize in discrete regions of the nucleoplasm. Thus, in AcMNPV infected cells a specific sorting of membranes occurs resulting in nuclear compartmentalization. Our observations show that some ODV envelope proteins associate at least transiently with cytoplasmic membranes located near the nucleus and with the nuclear envelope, including the INM. This association led us to consider two hypotheses: (i) transport of some ODV envelope proteins into the nucleus could be mediated through membranes, and (ii) regions of the INM could invaginate into the nucleus and function as a source of viral induced intranuclear microvesicles (6). Insect cells may contain cellular proteins with similarities to one or more of the viral envelope proteins; thus, the transport of viral proteins to the ODV envelope may represent a viral amplified, but normal, cellular pathway of integral membrane protein transport into the nuclear envelope (refs. 6 and 19 and unpublished results).
Transmembrane targeting or retention signals sequences function for specific integral membrane proteins to locate or retain proteins with distinct subset(s) of cellular membranes. In this study we demonstrated that a short sequence, composed predominantly of a hydrophobic domain derived from two ODV envelope proteins, is sufficient to direct proteins that are normally cytosolic (β-gal, ref. 20), or dispersed throughout the cell (GFP, refs. 16 and 17), into cytoplasmic membranes, the nuclear envelope, viral-induced intranuclear microvesicles, and the ODV envelope. This sequence apparently does not cause random insertion of proteins into cellular membranes in general, as the wild-type and fusion proteins are not detected on the plasma membrane (Fig. 2C) or budded virus envelope (5, 15). Wild-type ODV-E66, ODV-E25, and the fusion protein 23β-gal, when expressed under their native or ODV-E66 promoter, demonstrate a limited association with cytoplasmic membranes and the nuclear envelope. However, when these or similar fusion proteins are expressed at high levels under the control of the polyhedrin promoter, they clearly associate with cytoplasmic membranes that are closely associated with the nucleus and the outer and INM. We propose several models to explain these results.
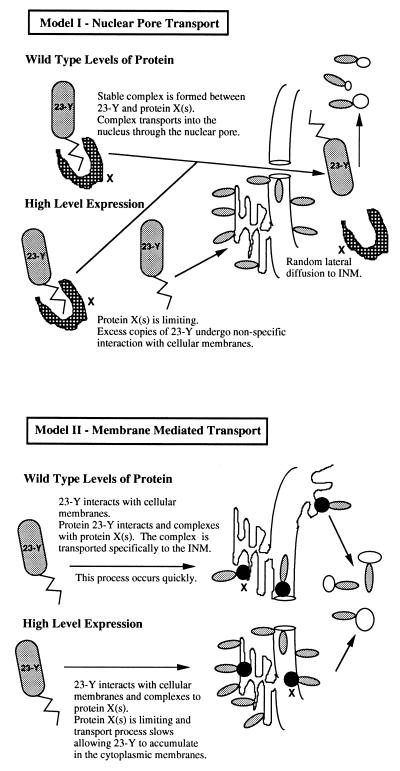
The most defined pathway of nuclear import is through the nuclear pore, however the viral 23/24 amino acid sequence and fusions thereof, do not contain a conventional nuclear targeting sequence. Fig. 4; model 1, illustrates that if other protein (X), binds to the N-terminal sequence (23-Y), it could contain a nuclear localization sequence and by itself, or complexed with additional proteins, serve to transport 23-Y into the nucleus through the nuclear pore. Upon entry, X, the binding/transport protein could disassociate, and release 23-Y to traffick to, and insert into membranes within the nucleus. If 23-Y is expressed at high levels, as would be the case with polyhedrin-promoter regulated expression, then protein X could become limiting and without the proper stoichiometry, 23-Y would randomly associate with cellular membranes. Several features argue against this model. (i) When expressed at high levels 23-Y is detected abundantly in both the outer and INM. This would suggest unregulated diffusion and/or retention from the outer to the INM. It would be difficult to explain how the INM could retain its structural and therefore, functional integrity separate from the ONM if such a random transport and/or retention mechanism is occurring. (ii) If 23-Y could randomly insert into any membrane when expressed at high levels, then it is difficult to understand why this protein is not detected at the plasma membrane.
A second model model predicts that 23-Y associates specifically with a subset of cellular membranes closely associated with the nuclear envelope, (presumably ER, or a subset thereof) where it then interacts with other protein(s), (X), which then functions to target or transport 23-Y from the outer to the INM (Fig. 4, model 2). If large amounts of 23-Y are incorporated into the cytoplasmic membrane (i.e., polyhedrin-promoter regulated expression), X is again limiting, and without the correct stoichiometry and efficient transport, 23-Y accumulates in the specific subset of cytoplasmic membranes (presumably ER).
Model 2 is more consistent with previous studies of protein transport to the nuclear envelope that indicate that cytoplasmic membranes, especially ER, may function as important intermediates in the transport pathway. LBR is first inserted into rough ER/ONM, then transported to the INM where it interacts with lamin B (8, 21). The integral nuclear envelope protein of avian erythrocytes, p18 is equally distributed between the ONM/rough ER and INM, and forms a complex with LBR and other proteins at the INM (22). N-glycoprotein gp210, an integral component of the nuclear pore complex, is first inserted into the rough ER/ONM where core N-glycosylation occurs, and then becomes incorporated into the nuclear pore complex (9) and, glycoprotein B of herpes simplex virus is first inserted into the rough ER/ONM where high-mannose N-glycans are added, and then transported to the INM in the high-mannose immature form (7, 23). Our results with 23GFP, 24GFP, and high levels of expression of ODV-E66 is also consistent with the ER functioning as an intermediate in the transport pathway to both the ONM and INM. Our data also describes an amino acid sequence that could interact with other protein(s) in this transport pathway. While these data are consistent with previous studies of hydrophobic domains, it also has unique differences. Studies using a hydrophobic domain of LBR to locate reporter protein fusions to the INM reveal that this domain cannot efficiently transport proteins >70 kDa into the INM. This led Soullam and Worman (24) to hypothesize that diffusion to the INM is coupled with nuclear pore interactions, and that a necessary feature of the transported proteins must be that they can fit through the lateral channels of the nuclear pore complex. This size restriction is not true for 23β-gal (≈120 kDa). It is unknown how membrane proteins are transported from the ER/ONM to the INM. This could be done utilizing the nuclear pore, or it is possible that a novel, as yet undescribed, pathway of protein movement analogous to other dual membrane transport systems may also exist.
While it is difficult to predict secondary structure of a hydrophobic domain, the hydrophobic sequences identified in this study may have some interesting differences as compared with other hydrophobic domains in INM proteins. nnpredict and saps analysis (25) predict that the 23 and 24 amino acid sequence ODV-E66 and ODV-E25 have a tendency to assume a β-sheet structure. This is in contrast to rat gp210, herpes simplex virus glycoprotein B segment 3, and chicken LBR (transmembrane segment 1), which are predicted to assume more of an α-helical orientation through a membrane bilayer. A single β-strand is believed to be unstable within a membrane, but would form stable complexes with other β-strands in the membrane. Such a structure has been proposed for ligand-gated ion channels and some neurotoxins (26–28). Thus, if the hydrophobic regions of ODV-E66 and ODV-E25 are associating with membranes via a β-strand they may be associating with themselves or other proteins to produce a stable complex. The hydrophobic domain of ODV-E66 and ODV-E25 is composed predominantly of the amino acids valine, isoleucine and leucines, amino acids that have been shown to play a strong role in stabilizing β sheets in a complex (29). The hydrophobic domains from gp210, herpes simplex virus glycoprotein B, and LBR are interrupted with proline, alanine, phenyalanine and threonines and these domains are located internally within the amino acid sequence. In contrast, the hydrophobic domains of ODV-E66 and ODV-E25 reside at the N terminus of each protein.
A novel feature of baculovirus infection is the induction of large amounts of intranuclear microvesicles and unit-membrane structures, some of which become the ODV envelope. We consider the results presented here to be consistent with the hypothesis that the INM may serve as a source of the intranuclear microvesicles. Localized and concentrated 23GFP and 24GFP fluorescence is first visualized closely associated with the nuclear envelope, both the outer and INM, and as the infection progresses, it moves to localized regions within the nucleus. As the pathway of trafficking and targeting of ODV envelope proteins is defined, we hope that this pathway will also reveal the source of membrane that becomes intranuclear microvesicles and the process involved in this induction and ultimately the source ODV envelope.
If AcMNPV is utilizing or redirecting the normal cellular pathways of membrane or membrane associated protein transport to the INM, then studies on baculovirus envelope proteins should reveal some details of the molecular biology and biochemistry utilized by cells for the regulation of these pathways. Investigations using these viral proteins as specific markers, may reveal some of the mechanisms and pathways utilized by cells to produce and retain the structural and functional integrity of the nuclear envelope.
Figure 5.
Models of protein transport into membranes within the nucleus.
Acknowledgments
We would like to thank Dr. Christian Oker-Blom (VTT Biotechnology and Food Research, Espoo, Finland) for providing the control recombinant virus pVL1393-GFP and Dr. G. Rohrmann (Oregon State University) for the generous gift of antisera to ODV-E25. This study was supported in part by National Institutes of Health Grant 2RO1GM47552 (M.D.S. and S.C.B.), Texas Agricultural Experiment Station Project TEXO8078 (M.D.S.), and Texas Higher Education Coordinating Board ARP/ATP Grant 999902–011 (S.C.B.).
ABBREVIATIONS
- AcMNPV
Autographa californica nuclear polyhedrosis virus
- ONM
outer nuclear membrane
- INM
inner nuclear membrane
- ODV
occlusion-derived virus
- ER
endoplasmic reticulum
- LBR
lamin B receptor
- β-gal
β-galactosidase, GFP, green fluorescent protein
References
- 1.Wiley D C, Skehel J J. In: Virology. Fields B N, Knipe D M, Chanock R M, Hirsch M S, Milnick J L, Monath T P, Roizman B, editors. New York: Raven; 1990. pp. 63–85. [Google Scholar]
- 2.Adams J R, McClintock J T. In: Atlas of Invertebrate Viruses. Adams J R, Bonami J R, editors. Boca Raton, FL: CRC; 1991. pp. 87–226. [Google Scholar]
- 3.Blissard G W, Rohrmann G F. Annu Rev Entomol. 1990;35:127–155. doi: 10.1146/annurev.en.35.010190.001015. [DOI] [PubMed] [Google Scholar]
- 4.Fraser M J. J Ultrastruct Mol Struct Res. 1986;95:189–195. [Google Scholar]
- 5.Hong T, Braunagel S C, Summers M D. Virology. 1994;204:210–222. doi: 10.1006/viro.1994.1525. [DOI] [PubMed] [Google Scholar]
- 6.Braunagel S C, Elton D M, Ma H, Summers M D. Virology. 1996;217:97–110. doi: 10.1006/viro.1996.0097. [DOI] [PubMed] [Google Scholar]
- 7.Gilbert R, Ghosh K, Rasile L, Ghosh H P. J Virol. 1994;68:2272–2285. doi: 10.1128/jvi.68.4.2272-2285.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Smith S, Blobel G. J Cell Biol. 1993;120:631–637. doi: 10.1083/jcb.120.3.631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wozniak R W, Blobel G. J Cell Biol. 1992;119:1441–1449. doi: 10.1083/jcb.119.6.1441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Webb N R, Summers M D. Technique (Philadelphia) 1990;2:173–188. [Google Scholar]
- 11.Summers, M. D. & Smith, G. E. (1987) Tex. Agric. Exp. Stn. Bull. 1555.
- 12.Sambrook J, Fritsch E F, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd Ed. Plainview, NY: Cold Spring Harbor Lab. Press; 1989. [Google Scholar]
- 13.Kabcenell A K, Atkinson P H. J Cell Biol. 1985;101:1270–1280. doi: 10.1083/jcb.101.4.1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braunagel S C, He H, Ramamurthy P, Summers M D. Virology. 1996;222:100–114. doi: 10.1006/viro.1996.0401. [DOI] [PubMed] [Google Scholar]
- 15.Russell R L Q, Rohrmann G F. Virology. 1993;195:532–540. doi: 10.1006/viro.1993.1404. [DOI] [PubMed] [Google Scholar]
- 16.Oker-Blom C, Orellana A, Keinanen K. FEBS Lett. 1996;389:238–243. doi: 10.1016/0014-5793(96)00593-5. [DOI] [PubMed] [Google Scholar]
- 17.Laukkanen M L, Oker-Blom C, Keinanen K. Biochem Biophys Res Commun. 1996;226:755–761. doi: 10.1006/bbrc.1996.1425. [DOI] [PubMed] [Google Scholar]
- 18.Summers M D, Arnott H J. J Ultrastruct Res. 1969;28:462–480. doi: 10.1016/s0022-5320(69)80034-1. [DOI] [PubMed] [Google Scholar]
- 19.Theilmann D A, Chantler J K, Stewart S, Flipsen H T M, Vlak J M, Crook N E. Virology. 1996;218:148–158. doi: 10.1006/viro.1996.0175. [DOI] [PubMed] [Google Scholar]
- 20.Jarvis D L, Bohlmeyer D A, Garcia A. Virology. 1991;185:795–810. doi: 10.1016/0042-6822(91)90551-l. [DOI] [PubMed] [Google Scholar]
- 21.Ye Q, Worman H J. J Biol Chem. 1994;269:11306–11311. [PubMed] [Google Scholar]
- 22.Simos G, Maison C, Georgatos S D. J Biol Chem. 1996;271:12617–12625. doi: 10.1074/jbc.271.21.12617. [DOI] [PubMed] [Google Scholar]
- 23.Torrisi M R, Lazzaro C D, Pavan A, Pereira L, Campadelli-Fiume G. J Virol. 1992;66:554–561. doi: 10.1128/jvi.66.1.554-561.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soullam B, Worman H J. J Cell Biol. 1995;130:15–27. doi: 10.1083/jcb.130.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Brendel V, Bucher P, Nourbakhsh I R, Blaisdell B E, Karlin S. Proc Natl Acad Sci USA. 1992;89:2002–2006. doi: 10.1073/pnas.89.6.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Shai Y. Trends Biochem Sci. 1995;20:460–464. doi: 10.1016/s0968-0004(00)89101-x. [DOI] [PubMed] [Google Scholar]
- 27.Fischbarg J, Cheung M, Li J, Iserovich P, Czegledy F, Kuang K, Garner M. Mol Cell Biochem. 1994;140:147–162. doi: 10.1007/BF00926753. [DOI] [PubMed] [Google Scholar]
- 28.Unwin, N. (1993) Cell 72/Neuron10, (Suppl.), 31–41.
- 29.Service R F. Science. 1996;274:34–35. doi: 10.1126/science.274.5284.34. [DOI] [PubMed] [Google Scholar]