Abstract
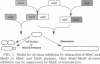
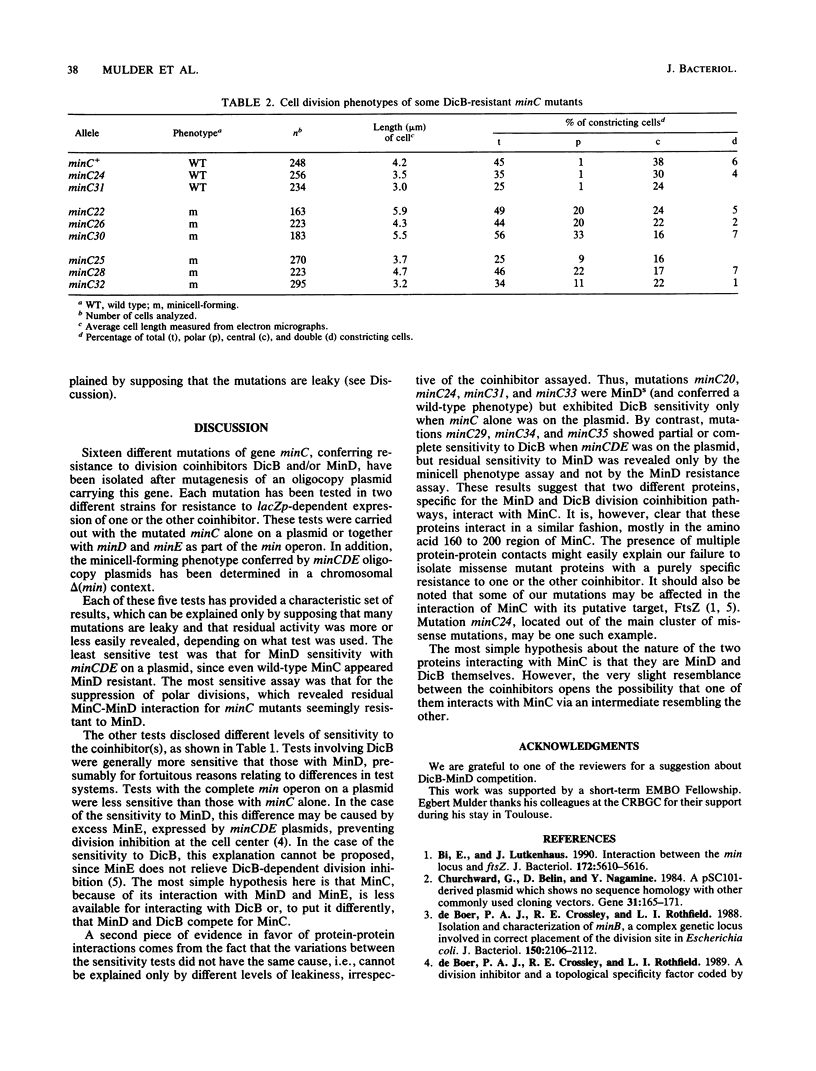
Proper positioning of division sites in Escherichia coli requires balanced expression of minC, minD, and minE gene products. Previous genetic analysis has shown that either MinD or an apparently unrelated protein, DicB, cooperates with MinC to inhibit division. We have isolated and sequenced minC mutations that suppress division inhibition caused by overproduction of either DicB or MinD proteins. Most missense mutations were located in the amino acid 160 to 200 region of MinC (231 amino acids). Some mutations exhibited preferential resistance to one or the other coinhibitor, suggesting that two distinct proteins, possibly MinD and DicB themselves, interact in slightly different manners with the same region of MinC to promote division inhibition.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bi E., Lutkenhaus J. Interaction between the min locus and ftsZ. J Bacteriol. 1990 Oct;172(10):5610–5616. doi: 10.1128/jb.172.10.5610-5616.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchward G., Belin D., Nagamine Y. A pSC101-derived plasmid which shows no sequence homology to other commonly used cloning vectors. Gene. 1984 Nov;31(1-3):165–171. doi: 10.1016/0378-1119(84)90207-5. [DOI] [PubMed] [Google Scholar]
- Ish-Horowicz D., Burke J. F. Rapid and efficient cosmid cloning. Nucleic Acids Res. 1981 Jul 10;9(13):2989–2998. doi: 10.1093/nar/9.13.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labie C., Bouché F., Bouché J. P. Isolation and mapping of Escherichia coli mutations conferring resistance to division inhibition protein DicB. J Bacteriol. 1989 Aug;171(8):4315–4319. doi: 10.1128/jb.171.8.4315-4319.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labie C., Bouché F., Bouché J. P. Minicell-forming mutants of Escherichia coli: suppression of both DicB- and MinD-dependent division inhibition by inactivation of the minC gene product. J Bacteriol. 1990 Oct;172(10):5852–5855. doi: 10.1128/jb.172.10.5852-5855.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mulder E., El'Bouhali M., Pas E., Woldringh C. L. The Escherichia coli minB mutation resembles gyrB in defective nucleoid segregation and decreased negative supercoiling of plasmids. Mol Gen Genet. 1990 Mar;221(1):87–93. doi: 10.1007/BF00280372. [DOI] [PubMed] [Google Scholar]
- Mulder E., Woldringh C. L. Actively replicating nucleoids influence positioning of division sites in Escherichia coli filaments forming cells lacking DNA. J Bacteriol. 1989 Aug;171(8):4303–4314. doi: 10.1128/jb.171.8.4303-4314.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud O., Mock M., Schwartz M. A technique for integrating any DNA fragment into the chromosome of Escherichia coli. Gene. 1984 Jul-Aug;29(1-2):231–241. doi: 10.1016/0378-1119(84)90183-5. [DOI] [PubMed] [Google Scholar]
- Rothfield L. I., DeBoer P., Cook W. R. Localization of septation sites. Res Microbiol. 1990 Jan;141(1):57–63. doi: 10.1016/0923-2508(90)90098-b. [DOI] [PubMed] [Google Scholar]
- Taschner P. E., Huls P. G., Pas E., Woldringh C. L. Division behavior and shape changes in isogenic ftsZ, ftsQ, ftsA, pbpB, and ftsE cell division mutants of Escherichia coli during temperature shift experiments. J Bacteriol. 1988 Apr;170(4):1533–1540. doi: 10.1128/jb.170.4.1533-1540.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teather R. M., Collins J. F., Donachie W. D. Quantal behavior of a diffusible factor which initiates septum formation at potential division sites in Escherichia coli. J Bacteriol. 1974 May;118(2):407–413. doi: 10.1128/jb.118.2.407-413.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woldringh C. L., Mulder E., Valkenburg J. A., Wientjes F. B., Zaritsky A., Nanninga N. Role of the nucleoid in the toporegulation of division. Res Microbiol. 1990 Jan;141(1):39–49. doi: 10.1016/0923-2508(90)90096-9. [DOI] [PubMed] [Google Scholar]
- de Boer P. A., Crossley R. E., Rothfield L. I. Central role for the Escherichia coli minC gene product in two different cell division-inhibition systems. Proc Natl Acad Sci U S A. 1990 Feb;87(3):1129–1133. doi: 10.1073/pnas.87.3.1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Boer P. A., Crossley R. E., Rothfield L. I. Isolation and properties of minB, a complex genetic locus involved in correct placement of the division site in Escherichia coli. J Bacteriol. 1988 May;170(5):2106–2112. doi: 10.1128/jb.170.5.2106-2112.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]