Abstract
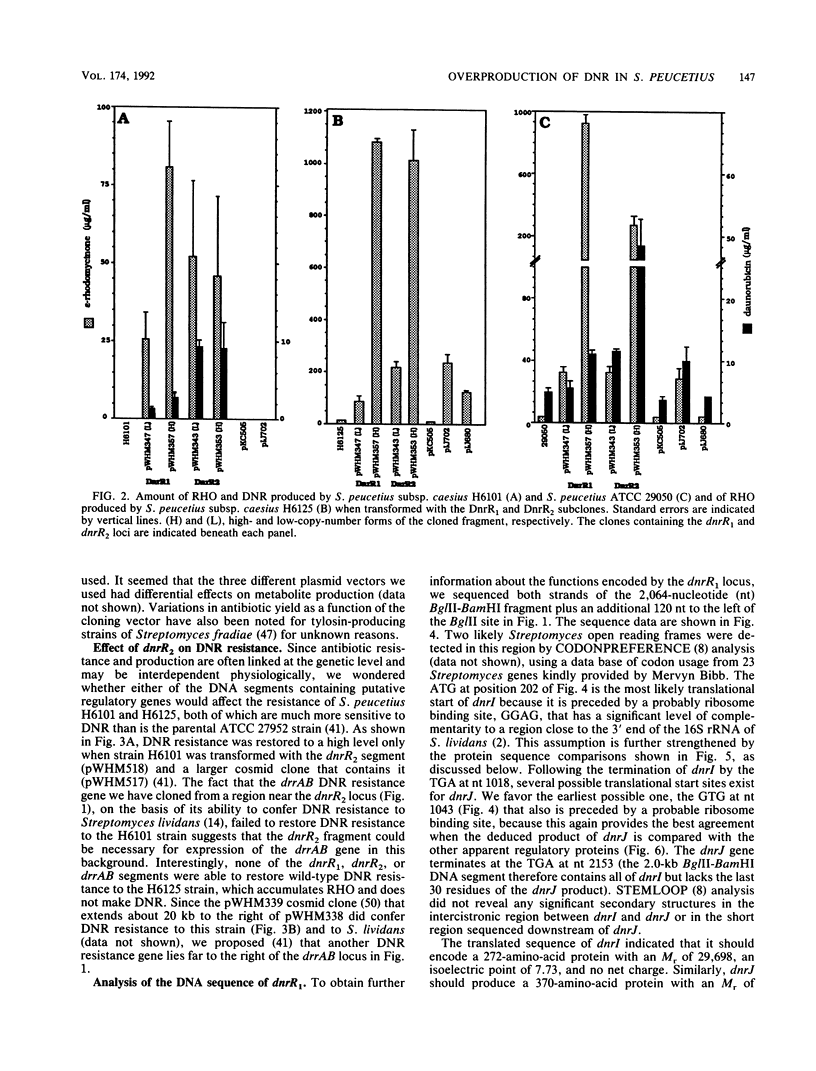
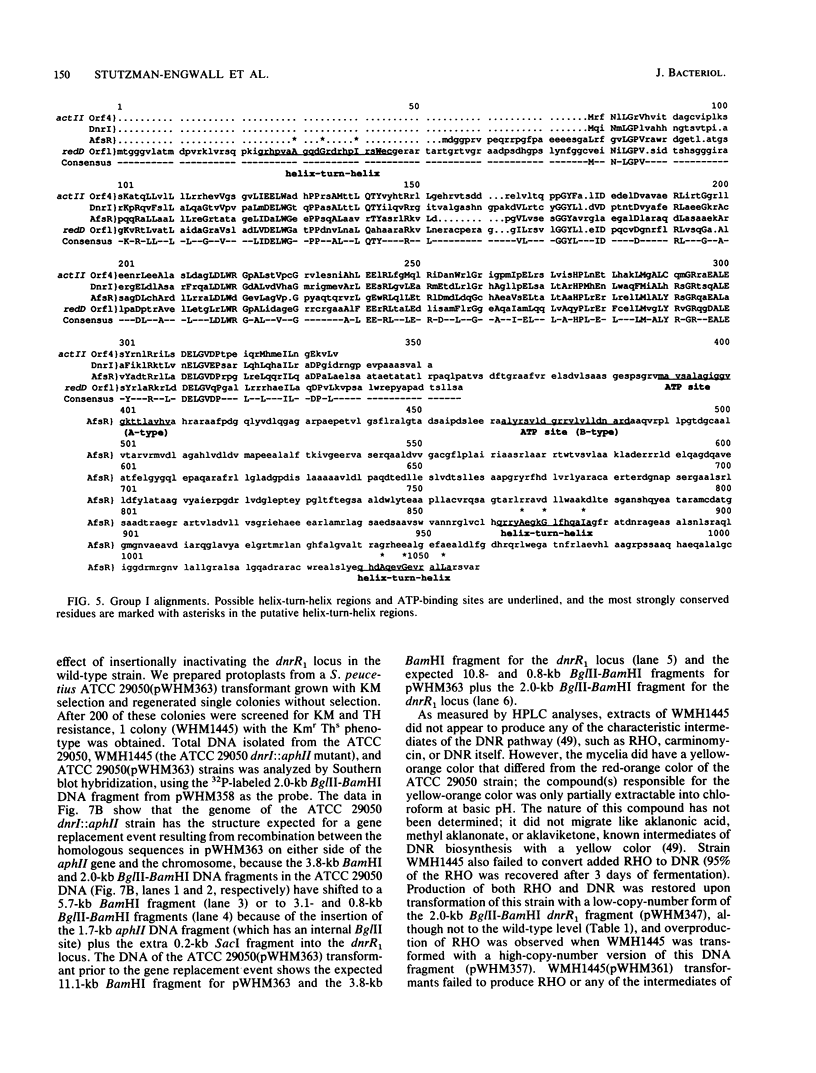
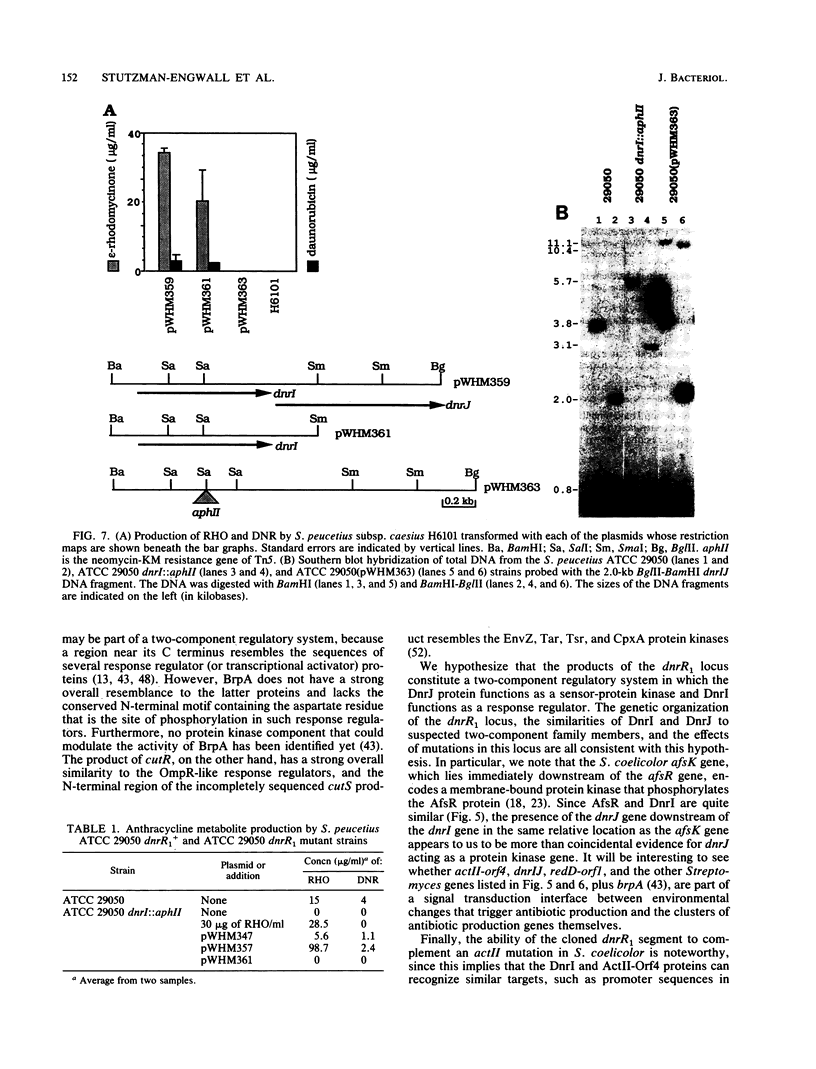
Two DNA segments, dnrR1 and dnrR2, from the Streptomyces peucetius ATCC 29050 genome were identified by their ability to stimulate secondary metabolite production and resistance. When introduced into the wild-type ATCC 29050 strain, the 2.0-kb dnrR1 segment caused a 10-fold overproduction of epsilon-rhodomycinone, a key intermediate of daunorubicin biosynthesis, whereas the 1.9-kb dnrR2 segment increased production of both epsilon-rhodomycinone and daunorubicin 10- and 2-fold, respectively. In addition, the dnrR2 segment restored high-level daunorubicin resistance to strain H6101, a daunorubicin-sensitive mutant of S. peucetius subsp. caesius ATCC 27952. Analysis of the sequence of the dnrR1 fragment revealed the presence of two closely situated open reading frames, dnrI and dnrJ, whose deduced products exhibit high similarity to the products of several other Streptomyces genes that have been implicated in the regulation of secondary metabolism. Insertional inactivation of dnrI in the ATCC 29050 strain with the Tn5 kanamycin resistance gene abolished epsilon-rhodomycinone and daunorubicin production and markedly decreased resistance to daunorubicin. Sequence comparison between the products of dnrIJ and the products of the Streptomyces coelicolor actII-orf4, afsR, and redD-orf1 genes and of the Streptomyces griseus strS, the Saccharopolyspora erythraea eryC1, and the Bacillus stearothermophilus degT genes reveals two families of putative regulatory genes. The members of the DegT, DnrJ, EryC1, and StrS family exhibit some of the features characteristic of the protein kinase (sensor) component of two-component regulatory systems from other bacteria (even though none of the sequences of these four proteins show a significant overall or regional similarity to such protein kinases) and have a consensus helix-turn-helix motif typical of DNA binding proteins. A helix-turn-helix motif is also present in two of the proteins of the other family, AfsR and RedD-Orf1. Both sets of Streptomyces proteins are likely to be trans-acting factors involved in regulating secondary metabolism.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anzai H., Murakami T., Imai S., Satoh A., Nagaoka K., Thompson C. J. Transcriptional regulation of bialaphos biosynthesis in Streptomyces hygroscopicus. J Bacteriol. 1987 Aug;169(8):3482–3488. doi: 10.1128/jb.169.8.3482-3488.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bibb M. J., Cohen S. N. Gene expression in Streptomyces: construction and application of promoter-probe plasmid vectors in Streptomyces lividans. Mol Gen Genet. 1982;187(2):265–277. doi: 10.1007/BF00331128. [DOI] [PubMed] [Google Scholar]
- Brennan R. G., Matthews B. W. The helix-turn-helix DNA binding motif. J Biol Chem. 1989 Feb 5;264(4):1903–1906. [PubMed] [Google Scholar]
- Chater K. F. Multilevel regulation of Streptomyces differentiation. Trends Genet. 1989 Nov;5(11):372–377. doi: 10.1016/0168-9525(89)90172-8. [DOI] [PubMed] [Google Scholar]
- Chater K. F. The improving prospects for yield increase by genetic engineering in antibiotic-producing Streptomycetes. Biotechnology (N Y) 1990 Feb;8(2):115–121. doi: 10.1038/nbt0290-115. [DOI] [PubMed] [Google Scholar]
- Devereux J., Haeberli P., Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhillon N., Hale R. S., Cortes J., Leadlay P. F. Molecular characterization of a gene from Saccharopolyspora erythraea (Streptomyces erythraeus) which is involved in erythromycin biosynthesis. Mol Microbiol. 1989 Oct;3(10):1405–1414. doi: 10.1111/j.1365-2958.1989.tb00123.x. [DOI] [PubMed] [Google Scholar]
- Distler J., Ebert A., Mansouri K., Pissowotzki K., Stockmann M., Piepersberg W. Gene cluster for streptomycin biosynthesis in Streptomyces griseus: nucleotide sequence of three genes and analysis of transcriptional activity. Nucleic Acids Res. 1987 Oct 12;15(19):8041–8056. doi: 10.1093/nar/15.19.8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feitelson J. S., Hopwood D. A. Cloning of a Streptomyces gene for an O-methyltransferase involved in antibiotic biosynthesis. Mol Gen Genet. 1983;190(3):394–398. doi: 10.1007/BF00331065. [DOI] [PubMed] [Google Scholar]
- Fernández-Moreno M. A., Caballero J. L., Hopwood D. A., Malpartida F. The act cluster contains regulatory and antibiotic export genes, direct targets for translational control by the bldA tRNA gene of Streptomyces. Cell. 1991 Aug 23;66(4):769–780. doi: 10.1016/0092-8674(91)90120-n. [DOI] [PubMed] [Google Scholar]
- Gross R., Aricò B., Rappuoli R. Families of bacterial signal-transducing proteins. Mol Microbiol. 1989 Nov;3(11):1661–1667. doi: 10.1111/j.1365-2958.1989.tb00152.x. [DOI] [PubMed] [Google Scholar]
- Guilfoile P. G., Hutchinson C. R. A bacterial analog of the mdr gene of mammalian tumor cells is present in Streptomyces peucetius, the producer of daunorubicin and doxorubicin. Proc Natl Acad Sci U S A. 1991 Oct 1;88(19):8553–8557. doi: 10.1073/pnas.88.19.8553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C., Aggarwal A. K. DNA recognition by proteins with the helix-turn-helix motif. Annu Rev Biochem. 1990;59:933–969. doi: 10.1146/annurev.bi.59.070190.004441. [DOI] [PubMed] [Google Scholar]
- Henikoff S. Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene. 1984 Jun;28(3):351–359. doi: 10.1016/0378-1119(84)90153-7. [DOI] [PubMed] [Google Scholar]
- Hong S. K., Kito M., Beppu T., Horinouchi S. Phosphorylation of the AfsR product, a global regulatory protein for secondary-metabolite formation in Streptomyces coelicolor A3(2). J Bacteriol. 1991 Apr;173(7):2311–2318. doi: 10.1128/jb.173.7.2311-2318.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopwood D. A. The Leeuwenhoek lecture, 1987. Towards an understanding of gene switching in Streptomyces, the basis of sporulation and antibiotic production. Proc R Soc Lond B Biol Sci. 1988 Nov 22;235(1279):121–138. doi: 10.1098/rspb.1988.0067. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Hara O., Beppu T. Cloning of a pleiotropic gene that positively controls biosynthesis of A-factor, actinorhodin, and prodigiosin in Streptomyces coelicolor A3(2) and Streptomyces lividans. J Bacteriol. 1983 Sep;155(3):1238–1248. doi: 10.1128/jb.155.3.1238-1248.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horinouchi S., Kito M., Nishiyama M., Furuya K., Hong S. K., Miyake K., Beppu T. Primary structure of AfsR, a global regulatory protein for secondary metabolite formation in Streptomyces coelicolor A3(2). Gene. 1990 Oct 30;95(1):49–56. doi: 10.1016/0378-1119(90)90412-k. [DOI] [PubMed] [Google Scholar]
- Horinouchi S., Suzuki H., Beppu T. Nucleotide sequence of afsB, a pleiotropic gene involved in secondary metabolism in Streptomyces coelicolor A3(2) and "Streptomyces lividans". J Bacteriol. 1986 Oct;168(1):257–269. doi: 10.1128/jb.168.1.257-269.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz E., Thompson C. J., Hopwood D. A. Cloning and expression of the tyrosinase gene from Streptomyces antibioticus in Streptomyces lividans. J Gen Microbiol. 1983 Sep;129(9):2703–2714. doi: 10.1099/00221287-129-9-2703. [DOI] [PubMed] [Google Scholar]
- Kieser T. Factors affecting the isolation of CCC DNA from Streptomyces lividans and Escherichia coli. Plasmid. 1984 Jul;12(1):19–36. doi: 10.1016/0147-619x(84)90063-5. [DOI] [PubMed] [Google Scholar]
- Kieser T., Hopwood D. A., Wright H. M., Thompson C. J. pIJ101, a multi-copy broad host-range Streptomyces plasmid: functional analysis and development of DNA cloning vectors. Mol Gen Genet. 1982;185(2):223–228. doi: 10.1007/BF00330791. [DOI] [PubMed] [Google Scholar]
- Kyte J., Doolittle R. F. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982 May 5;157(1):105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- Lawlor E. J., Baylis H. A., Chater K. F. Pleiotropic morphological and antibiotic deficiencies result from mutations in a gene encoding a tRNA-like product in Streptomyces coelicolor A3(2). Genes Dev. 1987 Dec;1(10):1305–1310. doi: 10.1101/gad.1.10.1305. [DOI] [PubMed] [Google Scholar]
- Malpartida F., Hopwood D. A. Molecular cloning of the whole biosynthetic pathway of a Streptomyces antibiotic and its expression in a heterologous host. 1984 May 31-Jun 6Nature. 309(5967):462–464. doi: 10.1038/309462a0. [DOI] [PubMed] [Google Scholar]
- Malpartida F., Hopwood D. A. Physical and genetic characterisation of the gene cluster for the antibiotic actinorhodin in Streptomyces coelicolor A3(2). Mol Gen Genet. 1986 Oct;205(1):66–73. doi: 10.1007/BF02428033. [DOI] [PubMed] [Google Scholar]
- Narva K. E., Feitelson J. S. Nucleotide sequence and transcriptional analysis of the redD locus of Streptomyces coelicolor A3(2). J Bacteriol. 1990 Jan;172(1):326–333. doi: 10.1128/jb.172.1.326-333.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Needleman S. B., Wunsch C. D. A general method applicable to the search for similarities in the amino acid sequence of two proteins. J Mol Biol. 1970 Mar;48(3):443–453. doi: 10.1016/0022-2836(70)90057-4. [DOI] [PubMed] [Google Scholar]
- Ohnuki T., Imanaka T., Aiba S. Self-cloning in Streptomyces griseus of an str gene cluster for streptomycin biosynthesis and streptomycin resistance. J Bacteriol. 1985 Oct;164(1):85–94. doi: 10.1128/jb.164.1.85-94.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otten S. L., Stutzman-Engwall K. J., Hutchinson C. R. Cloning and expression of daunorubicin biosynthesis genes from Streptomyces peucetius and S. peucetius subsp. caesius. J Bacteriol. 1990 Jun;172(6):3427–3434. doi: 10.1128/jb.172.6.3427-3434.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raibaud A., Zalacain M., Holt T. G., Tizard R., Thompson C. J. Nucleotide sequence analysis reveals linked N-acetyl hydrolase, thioesterase, transport, and regulatory genes encoded by the bialaphos biosynthetic gene cluster of Streptomyces hygroscopicus. J Bacteriol. 1991 Jul;173(14):4454–4463. doi: 10.1128/jb.173.14.4454-4463.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richardson M. A., Kuhstoss S., Solenberg P., Schaus N. A., Rao R. N. A new shuttle cosmid vector, pKC505, for streptomycetes: its use in the cloning of three different spiramycin-resistance genes from a Streptomyces ambofaciens library. Gene. 1987;61(3):231–241. doi: 10.1016/0378-1119(87)90187-9. [DOI] [PubMed] [Google Scholar]
- Rudd B. A., Hopwood D. A. Genetics of actinorhodin biosynthesis by Streptomyces coelicolor A3(2). J Gen Microbiol. 1979 Sep;114(1):35–43. doi: 10.1099/00221287-114-1-35. [DOI] [PubMed] [Google Scholar]
- Stock J. B., Ninfa A. J., Stock A. M. Protein phosphorylation and regulation of adaptive responses in bacteria. Microbiol Rev. 1989 Dec;53(4):450–490. doi: 10.1128/mr.53.4.450-490.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stutzman-Engwall K. J., Hutchinson C. R. Multigene families for anthracycline antibiotic production in Streptomyces peucetius. Proc Natl Acad Sci U S A. 1989 May;86(9):3135–3139. doi: 10.1073/pnas.86.9.3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M., Takada H., Imanaka T. Nucleotide sequence and cloning in Bacillus subtilis of the Bacillus stearothermophilus pleiotropic regulatory gene degT. J Bacteriol. 1990 Jan;172(1):411–418. doi: 10.1128/jb.172.1.411-418.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng H. C., Chen C. W. A cloned ompR-like gene of Streptomyces lividans 66 suppresses defective melC1, a putative copper-transfer gene. Mol Microbiol. 1991 May;5(5):1187–1196. doi: 10.1111/j.1365-2958.1991.tb01892.x. [DOI] [PubMed] [Google Scholar]
- Vara J., Lewandowska-Skarbek M., Wang Y. G., Donadio S., Hutchinson C. R. Cloning of genes governing the deoxysugar portion of the erythromycin biosynthesis pathway in Saccharopolyspora erythraea (Streptomyces erythreus). J Bacteriol. 1989 Nov;171(11):5872–5881. doi: 10.1128/jb.171.11.5872-5881.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward J. M., Janssen G. R., Kieser T., Bibb M. J., Buttner M. J., Bibb M. J. Construction and characterisation of a series of multi-copy promoter-probe plasmid vectors for Streptomyces using the aminoglycoside phosphotransferase gene from Tn5 as indicator. Mol Gen Genet. 1986 Jun;203(3):468–478. doi: 10.1007/BF00422072. [DOI] [PubMed] [Google Scholar]
- Yanisch-Perron C., Vieira J., Messing J. Improved M13 phage cloning vectors and host strains: nucleotide sequences of the M13mp18 and pUC19 vectors. Gene. 1985;33(1):103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- Yoshimoto A., Matsuzawa Y., Oki T., Takeuchi T., Umezawa H. New anthracycline metabolites from mutant strains of Streptomyces galilaeus MA144-M1. I. Isolation and characterization of various blocked mutants. J Antibiot (Tokyo) 1981 Aug;34(8):951–958. doi: 10.7164/antibiotics.34.951. [DOI] [PubMed] [Google Scholar]




