Abstract
A model that explains the stoichiometric packaging of the chromosomes of Φ6, a bacteriophage with a genome of three unique double-stranded RNA segments, is proposed and supported. Ordered switches in packaging specificity and RNA synthesis are determined by the amount of RNA within the procapsid. The plus strand of segment S binds to one of several sites on the outside of the empty procapsid. The RNA enters and the procapsid expands so that the S sites are lost and M sites appear. Packaging of segment M results in the loss of the M sites and the appearance of the L sites. Packaging of L readies the particle for minus-strand synthesis. If any of the segments is less than normal size, packaging of that class of segments continues until the normal content of RNA for that segment is packaged and the binding sites then change.
Keywords: genomic packaging, Reoviridae
Most plus-strand RNA viruses contain one or two molecules of single-stranded RNA, and it is believed that the capsid assembles around the RNA (1). Bacteriophages with double-stranded DNA genomes package their DNA into preformed proheads (1). The well-studied viral systems that use preformed procapsids for packaging are the double-stranded DNA bacteriophages such as λ, T4, T7, P22, φ29, and the single-stranded DNA phage φX174 (1–3). In these cases, only one segment is packaged, and the packaging specificity is determined by a set of proteins that are not permanent components of the procapsid, but are specifically involved in packaging and, in some cases, in cutting of a concatameric precursor of the viral genome. In some cases, the packaging proceeds until a “headful” is obtained, in others a specific genomic unit is packaged. All of these systems depend upon NTP hydrolysis for packaging, although the φX174 system is coupled to DNA synthesis (3). In the case of the double-stranded DNA phages, there is a unique portal for entry and exit of the DNA. In the case of φX174, there is no clear portal structure (3) .
Several classes of virus carry a larger number of unique chromosomes within the same particle. The reoviridae contain 10, 11, or 12 double-stranded RNA chromosomes in each virion (4), and there is a consensus that the genomic content of each particle is uniform and stoichiometric (5). Influenza virus contains about 12 single-stranded RNA minus-strand segments per particle, but the complete complement is 8. The packaging of flu RNA is believed to be random (6), but there is some support for specific packaging (7). Φ6 is the only known bacteriophage that contains double-stranded RNA (8). It has three unique chromosomes per particle and the packaging is very precise (9). In this paper we propose a model to explain stoichiometric packaging of the Φ6 genome.
We have shown that preformed procapsids are capable of packaging the plus strands of the Φ6 chromosomes in vitro (10). The packaging is dependent upon nucleoside triphosphate (11) and is independent of minus-strand synthesis. Packaging is serially dependent (12, 13), since the plus strand of segment S can package alone, but M is packaged only after S, and segment L is packaged only after M. A sequence of about 200 nucleotides near the 5′ end of each plus strand is necessary and sufficient for packaging when in combination with a sequence of 18 nucleotides at the 5′ end that is essentially identical for each of the three segments (14). The 200-base pac sequence is completely different for each segment, and small changes destroy packaging competence.
An extensively deleted RNA molecule containing the pac sequence can still be packaged and can serve as a template for minus-strand synthesis (14, 15). Molecules lacking the normal 3′ ends can be packaged but do not serve as templates for minus-strand synthesis. They do, however, promote the packaging of the next class of segments in the series S, M, L (15). Curiously, molecules smaller than normal are packaged in multiples so that the total amount of RNA packaged per segment class is roughly equal to the normal size of that segment (16); a molecule that is one-fifth normal size is packaged in about five copies. This was shown in both in vitro packaging experiments and in the in vivo construction of viable phage with more than three molecules of RNA (17) .
The procapsid of Φ6 is composed of four proteins: P1, P2, P4, and P7 (18). P1 and P4 are present in 120 copies, P2 in about 12, and P7 in about 100 (9). P2 has the sequence motifs of viral RNA polymerase (19). P4 has NTPase activity that is the same as the NTP specificity for RNA packaging (20, 21). Procapsids cannot be made without P1, and those missing P4 or P7 are severely defective in packaging (10, 22). Procapsids missing P2 are somewhat defective in packaging, but are completely defective in RNA synthesis (22, 23). Empty procapsids of Φ6 have the appearance of a compressed dodecahedron (10). Three-dimensional reconstructions from cryoelectron micrographs of procapsids have clearly shown this type of structure (S. Butcher, T. Dokland, S. Fuller, P. Ojala, and D. Bamford, personal communication).
The Φ6 procapsid has no apparent unique portal vertex. A study with RNA molecules containing hairpin structures indicated that the portals are small and that there is more than one per procapsid (24). It is tempting to think that each of the 12 faces of the procapsid might contain an entry port. How then, do we reconcile a symmetrical structure which packages three different RNA molecules in a specific order, so that the amount of RNA packaged per segment type is roughly constant?
MATERIALS AND METHODS
Bacterial Strains and Plasmids.
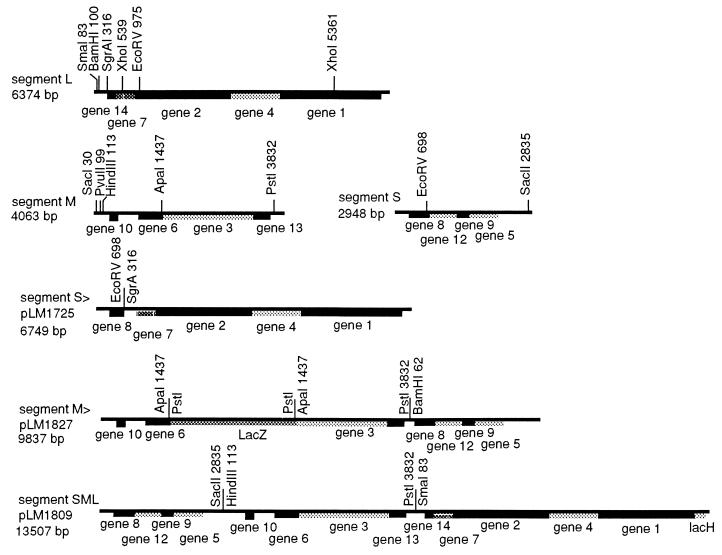
Escherichia coli strain JM109 (25) was used for the propagation of all plasmids. Plasmid pLM450 contains a cDNA copy of genomic segment L of the virus Φ6, encoding the four procapsid proteins (26). Plasmids pLM659, pLM656, and pLM682 contain cDNA copies of the genomic segments S, M, and L, respectively, in the pT7T3 19U vector (27). Plasmid pLM1157 contains a cDNA copy of segment L with the deletion of the XhoI fragment from positions 545 to 5366. Its transcript is about 1.5 kb and is packaged more efficiently than the normal L segment in vitro. Plasmids that produce transcripts of various sizes were constructed by using standard techniques (28) and are shown in Fig. 1.
Figure 1.
Restriction maps of the cDNA copies of the genomic segments of Φ6, L, M, and S as well as diagrams of the chimeric constructions used in this study. The sequences are all embedded in plasmid pT7T319U and the transcripts produced by T7 RNA polymerase have the correct base sequence at the 5′ ends. pLM1725 was prepared by ligating the 5′ end of segment S cut at the EcoRV site to a blunted SgrAI site of segment L. The aim was to prepare a transcript with the pac site of S, containing no sequence from M, but having a size close to that of the sum of S and M. pLM1827 was prepared by ligating segment M cut at the PstI site to segment S cut at the BamHI site to form an MS chimera (pLM1822) and then inserting the complete lacZ gene from plasmid 1871 into the ApaI site of M with PstI linkers. The aim was to prepare a transcript with the pac site of M, containing no sequence from L, but having a size close to that of the sum of M and L. pLM1809 contains the sequence of the three segments in the order S, M, L with the addition of a 427-base lacZα fragment to attain the size of the complete genome. The aim was to prepare a transcript with the pac site of S that was of the same size as the total Φ6 genome.
In Vitro Synthesis of Radioactive Plus Sense Transcripts by T7 Polymerase.
Plasmids were cut with restriction endonuclease XbaI. The resulting 5′ overhang was removed with Mung bean nuclease before transcription with T7 RNA polymerase (27). This procedure generated 3′ ends of the transcripts identical to that of the Φ6 virus-produced message RNA. The polymerase reaction contained 2 mM each of UTP, ATP, GTP, and CTP and 400 microcuries of [α-32P]UTP per ml. The RNA was purified by filtering through G-50 Sephadex spin columns (Boehringer Mannheim).
Packaging Reaction Conditions.
Frozen purified procapsid preparations produced in E. coli JM109 carrying plasmid pLM450 (10) were thawed and incubated for 90 min at 28°C in a 12.5-μl packaging reaction (12) containing about 150 ng of [32P]UTP-labeled single-stranded Φ6 RNA for each segment. Approximately 1 μg of procapsid was used per reaction. The samples were then treated with 10 units of RNase I (RNase One; Promega) (29) and incubated for 30 min at 28°C. Ten microliters stop solution [3× sample buffer (30)/1 μg carrier RNA/25 mM EDTA] was added and the samples heated at 85°C for 5 min. The samples were then electrophoresed in 1% or 1.5% agarose gels.
Minus-Strand Synthesis.
Conditions for minus-strand synthesis were similar to those reported previously (11, 12, 15). Equimolar amounts of each segment were use an the total RNA concentration was about 100 μg/ml in a 25 μl reaction volume. The reaction was stopped by adding 3× sample buffer (30) and EDTA to final concentrations of 10 mM. The reaction products were analyzed with 1 μg of carrier RNA on 1.5% agarose gels containing 0.1% SDS in 0.5× TBE buffer (28) that were subsequently dried and the 32P-labeled RNA was visualized after autoradiography with a Cronex (DuPont) enhancing screen. Plus-strand synthesis was performed similarly to minus-strand synthesis, but 1 mM MnCl2 was added to the reaction and ammonium acetate was 80 mM instead of 100 mM.
RESULTS AND DISCUSSION
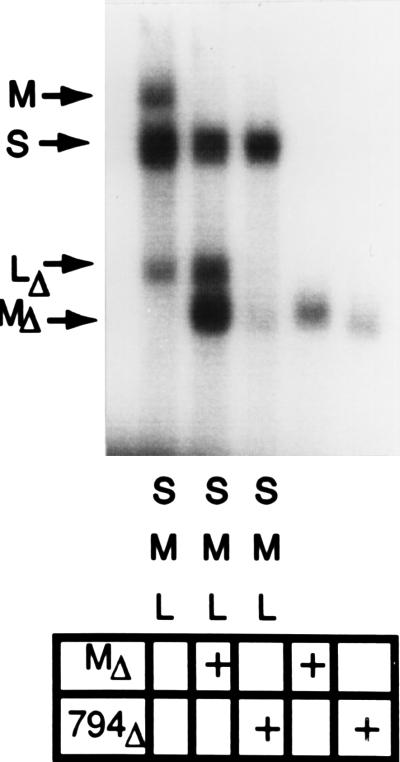
The first question we pose is: “Where does the specific binding of the RNA segments take place?” Juuti and Bamford (22) have shown that there is binding activity on the outside of procapsids, but that the specificity is for Φ6 RNA in general, not for specific segments. We found that direct binding experiments did not demonstrate segment specificity and therefore devised an alternative approach. Earlier experiments had shown that there are a number of dispensable nucleotides between the 5′ 18-base consensus sequence and the 200-base packaging sequence of segment M (14). Deletions in this region do not compromise packaging; a deletion of nucleotides 17–43 did not jeopardize packaging or minus-strand synthesis. However, a deletion of nucleotides 11–43 (pLM794), which removes 7 of the 18 consensus bases, was not packaged. We then determined whether this RNA would compete with normal mRNA for packaging. In Fig. 2 we see that a mixture of radioactive segments S, M, and L are packaged and protected from RNase. Note that the segment L used was smaller than the others since packaging of normal size L is inefficient in vitro. When a 20-fold excess of radioactive segment M, truncated at position 1192, was added, the packaging of radioactive normal sized M was diminished but that of the other segments was essentially unchanged. When a 20-fold excess of the truncated M transcript of pLM794, which lacks bases 11–43, was added, the packaging of segment S was normal, the packaging of segment M was diminished as before, but that of segment L was also diminished, indicating that the competitor RNA did not replace normal segment M. The competitor RNA was not packaged. There was a small amount of protected RNA when the 20-fold excess of normal or deletion RNA was tested in the absence of the other segments. This small amount of protection was probably due to abnormal packaging (see ref. 12). This experiment was repeated with normal-sized nonradioactive segments M and with the pLM794 deletion as competitors. The nonradioactive segment with the normal M pac site competed with radioactive M only, while the pLM794 RNA competed with both M and L (results not shown).
Figure 2.
Autoradiogram of gel analysis. In vitro packaging of labeled plus strands showing competition for packaging by RNA that is not packaged. Equal amounts of 32P-labeled transcripts of plasmids pLM658 and pLM656 were used for segments S and M, respectively. Plasmid pLM1157, which yields a 1.5-kb transcript, was used for segment L because it packages better than normal segment L. The packaging of the three segments is illustrated in lane 1, which shows an autoradiogram of an agarose gel analysis of a packaging mixture that had been treated with RNase I to degrade any unpackaged RNA. The conditions of packaging were as described (12). The effects of competition on packaging are shown in lanes 2 and 3, where a 20-fold excess of competitor RNA is added. In lane 2 the RNA is a normal segment M truncated at the SalI site at 1192. In lane 3 the RNA is derived from plasmid 794 also truncated at 1192. Plasmid pLM794 produces a transcript with a deletion of nucleotides 11–43. In lanes 4 and 5 the packaging of the competitor RNAs are shown in the absence of the normal plus strands.
To support further the idea of packaging competition for sites outside the procapsid, we prepared new transcripts that would compete for packaging without themselves being packaged. First, deleted templates were prepared with nucleotides 11–18 in segments S, L, and M deleted. The plus-strand transcripts of these constructs were packaged by procapsids and supported packaging of other segments. This result indicated that the loss of nucleotides 11–18 did not account for the lack of packaging of the transcript of pLM794. It appears rather that the spacing between the 18-base identity element and the pac sequence is critical. We then prepared a set of templates for segment S that contained deletions of nucleotides 11–23, 11–32, and 11–43. The plus strands of S with the deletions of nucleotides 11–18 or 11–23 were packaged and facilitated the packaging of M and L. The plus strand of S with a deletion from nucleotides 11–43 was not packaged, nor did it compete for the packaging of normal S plus strands. This segment also failed to support or interfere with the packaging of M and L. The plus strand of S with the deletion of nucleotides 11–32 was not packaged, but competed with the packaging of normal S and prevented the packaging of M and L (results not shown). These observations complemented those obtained with the plus strand of segment M that was missing nucleotides 11–43. In both cases, an RNA molecule that is not packaged by the procapsid competes for packaging with its normal homologue and fails to promote packaging of those plus strands that are usually dependent upon them. Molecules with larger deletions do not compete and are not packaged; molecules with smaller deletions are packaged normally, compete for packaging, and promote the packaging of dependent molecules.
Similar experiments were carried out with RNA totally missing the 18-base consensus sequence. These RNA molecules did not compete with normal packaging even at a 20-fold excess. This result was obtained for segments M and L. Another experiment tested the ability of RNA with deletions in the packaging sequence to compete with normal segments in M packaging (14). These RNA molecules also failed to compete at a 20-fold excess.
These experiments show that packaging can be competitively inhibited by an RNA that is not packaged, supporting the idea that the segment-specific binding sites are on the outside of the procapsid. The 200-base internal packaging sequence alone cannot compete with the packaging of normal RNA. At least some of the 18-base terminal sequence is needed for competition and the distance between the 200-base packaging sequence and the 5′ terminus seems to be important.
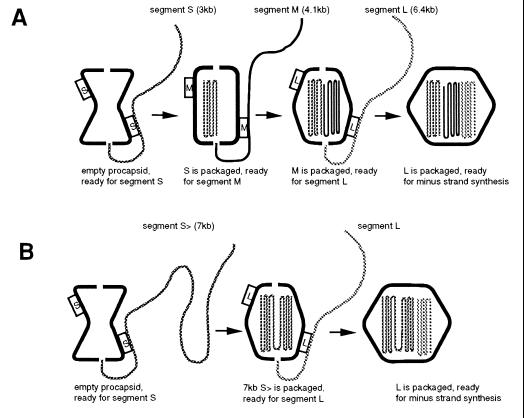
The Model.
The procapsid is in a compressed form after initial assembly (10). We propose that there are at least 12 binding sites for segment S on the dodecahedral procapsid, at least 1 per face. We propose that segment S first binds to one of these sites (Fig. 3A), positioning the 18-base 5′ sequence at the nearby entrance port. We propose further that the NTPase P4 protein (20), necessary to draw the RNA into the procapsid, is located at this port. The association of the RNA at the binding site is weak and therefore does not prevent subsequent 5′ to 3′ packaging (24). As segment S is packaged, the procapsid expands or changes conformation so that the S binding sites disappear, and the M binding sites appear. These may involve the same or different parts of P1. The process of expansion may resemble that of prohead expansion seen in double-stranded DNA bacteriophages (31, 32). Segment M then binds to the new site, its 5′ end is positioned at the nearby entry port and uptake commences. The M RNA might be binding to the same face as did S, or to another face. As the M RNA is packaged, the M-specific binding sites are lost and binding sites for L appear. Segment L binds and is packaged, and the particle again expands to prepare for minus-strand synthesis.
Figure 3.
(A) The packaging model. The procapsid shows only binding sites for S at the beginning. After a full size S is packaged, the S sites disappear and M sites appear. After a full size M is packaged, the M sites disappear and L sites appear. After a full size L is packaged, minus-strand synthesis commences. After minus-strand synthesis is completed, plus-strand synthesis commences. (B) If segment S is of the size equal to the sum of both S and M, the S sites will disappear and the L sites will appear and segment L will be packaged without segment M.
If any of the segments is truncated at its 3′ end or has internal deletions not directly affecting packaging, then the packaging of one molecule might not be sufficient to cause the necessary expansion. The procapsid would therefore remain competent to package another molecule of the same specificity class. In this case a procapsid could package n molecules of a given segment if its size was 1/n of the normal size. It could not package a molecule of another specificity class to make up the mass deficit because the packaging specificity of the procapsid would not have changed.
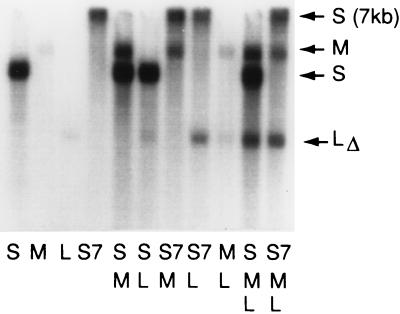
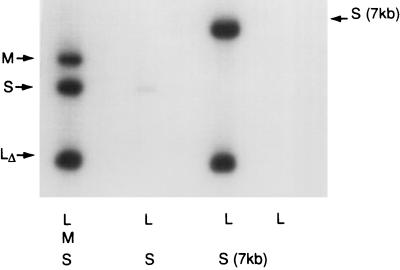
The model predicts that an RNA molecule with the packaging sequence of S but a size close to that of the sum of S and M (7 kb) could promote the packaging of segment L (Fig. 3B). In Fig. 4 we show that a molecule of 6.7 kb, containing no M sequence, does promote the packaging of segment L. In fact, this chimeric molecule is mostly derived from segment L, but lacking its pac sequence (Fig. 1). In Fig. 5, we show that the 6.7-kb S segment plus segment L supports minus-strand synthesis. These experiments indicate that the dependence of L on M is not a function of M itself, but simply depends on the amount of RNA that has been packaged.
Figure 4.
In vitro packaging of labeled plus strands showing that segment L can be packaged in the absence of M if segment S is made larger. The RNA in the packaging mixtures is shown below each lane. Segment S is derived from pLM658, segment M from 656, and segment L from pLM1157 as in Fig. 2. Segment S7 is the transcript of pLM1725 (Fig. 1) which is 6.8 kb. Note that segment S packages alone, whereas M and L package poorly alone. Segment M packages well in the presence of S and segment L packages well in the presence of S and M. Note that segment S7 packages alone (lane 4) and that segment L packages with segment S7 (lane 8) but not well with either S alone or M alone.
Figure 5.
Minus-strand synthesis showing that segment L can be packaged in the presence of segment S7 and turn on minus-strand synthesis. Unlabeled plus-strand RNA was added to procapsids in the presence of labeled NTPs as described (12). Minus-strand synthesis normally occurs when all three segments are packaged. Segment S does not support packaging or minus-strand synthesis of segment L, but S7 does.
The synthesis of the complementary minus strands of the packaged plus strands normally depends on the packaging of all three genomic segments (15, 33). Under special conditions minus-strand synthesis can be turned on by any combination of segments containing segment L, leading to the conclusion that segment L is the determinant for minus-strand synthesis (34). It has also been shown that plus-strand synthesis, in turn, depends on the completion of minus-strand synthesis on the segment L template (15, 34). In contrast, the results presented below show that the signals for the onset of both minus-strand and plus-strand synthesis require only that a critical amount of RNA be packaged, independent of the presence of segment L or sequence fragments of segment L.
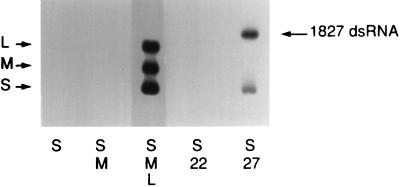
A chimeric segment was prepared that contains the normal sequence of segment M at the 5′ end, but is extended so as to have the size of the sum of both M and L. It contains a copy of M as well as a copy of segment S and the sequence of the complete lacZ gene. The transcript of this construct, pLM1827, was 9.8 kb and contained no sequences derived from segment L (Fig. 1). The plus-strand transcript of this construct could be packaged by normal procapsids if segment S was packaged at the same time. Minus-strand synthesis was observed in this case, although minus-strand synthesis did not occur if normal segments S and M were packaged without segment L (Fig. 6). Under conditions where plus-strand synthesis could be observed, the same construct resulted in plus-strand synthesis as well (Fig. 7). These results extend the model to encompass not only the order and amount of packaging of the genomic segments, but the switching on of both minus- and plus-strand synthesis. The conclusion is that the amount of RNA packaged or synthesized at various stages is sufficient to modify the behavior of the procapsid so as to bring it to the next step of reactivity. Finally, a plasmid, pLM1809, was constructed that produced a transcript containing segments S, M, and L joined together with the pac sequence of only segment S (Fig. 1). This transcript, which consists of 13.5 kb, the size of the total Φ6 genome, could be packaged by normal procapsids and led to both minus-strand and plus-strand synthesis (Fig. 7).
Figure 6.
Minus-strand synthesis without segment L. Unlabeled RNA was added to procapsids along with labeled NTPs as described (12). Segment S alone or S and M together do not turn on minus-strand synthesis, but S, M, and L together do promote synthesis. Segment S and the transcript of pLM1827 (Fig. 1) do turn on minus-strand synthesis although the latter transcript contains no sequence of segment L. The pLM1827 transcript is 9.8 kb, which is close to the sum of M plus L, 10.5 kb. The transcript of pLM1822, which is 6.8 kb, does not turn on minus-strand synthesis. Lane 3, which has RNA of S, M, and L, is exposed for one-tenth the time of the other lanes. dsRNA, double-stranded RNA.
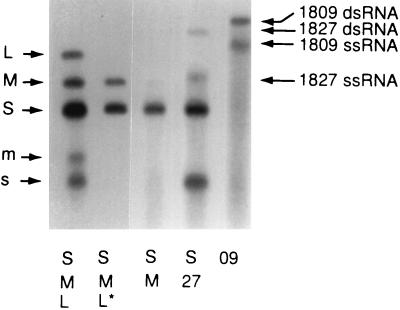
Figure 7.
Plus-strand synthesis without segment L. Unlabeled RNA was added to procapsids along with labeled NTPs as described (12), but with 1 mM Mn2+ in addition to the 3 mM Mg2+ and 80 mM NH4 acetate instead of 100 mM to promote plus-strand synthesis. In lane 1, segments L, M, and S served as templates for minus-strand synthesis (L, M, S) and for plus-strand synthesis (m, s). The plus strand of segment L is obscured by S. In lane 2 minus-strand synthesis occurs in the presence of intact segments S and M when packaged with segment L truncated at EcoRV (Fig. 1). In this case plus-strand synthesis does not occur as noted previously. Minus-strand synthesis is observed in lane 3 because Mn allows minus-strand synthesis without complete packaging (see Fig. 6). Plus-strand synthesis is seen in lane 4 for both segment S and the transcript of pLM1827, even though this transcript has no sequences of L. In lane 5 minus- and plus-strand synthesis is seen for the transcript of pLM1809 (Fig. 1), which has the pac site of S and the entire genetic complement of Φ6 and a size of 13.5 kb. Lanes 1 and 2 were exposed for one-tenth the time of the other lanes. dsRNA, double-stranded RNA; ssRNA, single-stranded RNA.
Is the Model Applicable to the Reoviridae?
The mechanism of packaging the 10, 11, or 12 genomic segments of reovirus, rotavirus, orbivirus, and wound tumor virus is not known. Although assembled cores of rotavirus are capable of minus-strand synthesis on rotavirus plus-strand templates, in vitro packaging has not been attained (35). In order for the present model to work for the Reoviridae there would have to be 10, 11, or 12 distinct conformational changes in the viral cores. This seems unlikely, but no alternative model has been proposed that is simpler or more appealing. Chromosomal rearrangements have been found in the rotavirus, but these seem to involve changes that increase the size of individual segments (36). Segments with internal deletions have been found in wound tumor virus (37) and in reovirus (38), but in these cases, a mixture of normal and small segments is packaged, making it difficult to determine the actual stoichiometry of the genomes. It is also possible that the number of double-stranded RNA genomic segments in a particle is limited by the number of molecules of polymerase in the particle and that number seems likely to be 12. In the case of Φ6, which normally has only three segments, there is leeway for dealing with multiple copies of shorter segments, but in the case of the Reoviridae, there may be additional constraints because of the high number of segments normally packaged. It would be of great interest to develop a complementation system for the Reoviridae that would enable isolation and propagation of virus constructs containing large deletions in particular segments.
Acknowledgments
We thank David Dubnau for reading the manuscript. This work was supported by Grant GM34352 from the National Institutes of Health to L.M.
References
- 1.Casjens S. In: Virus Structure and Assembly. Casjens S, editor. Boston: Jones & Bartlett; 1985. pp. 75–147. [Google Scholar]
- 2.Black L W. In: The Bacteriophages. Calendar R, editor. Vol. 2. New York: Plenum; 1988. pp. 321–373. [Google Scholar]
- 3.Aoyama A, Hamatake M, Hayashi M. Proc Natl Acad Sci USA. 1981;78:7285–7289. doi: 10.1073/pnas.78.12.7285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tyler K L, Fields B N. In: Virology. Fields B N, Knipe D M, editors. New York: Raven; 1990. pp. 1271–1273. [Google Scholar]
- 5.Spendlove R S, McClain M E, Lennette E H. J Gen Virol. 1970;8:83–93. doi: 10.1099/0022-1317-8-2-83. [DOI] [PubMed] [Google Scholar]
- 6.Enami M, Sharma G, Benham C, Palese P. Virology. 1991;185:291–298. doi: 10.1016/0042-6822(91)90776-8. [DOI] [PubMed] [Google Scholar]
- 7.Duhaut S D, McCauley J W. Virology. 1996;216:326–337. doi: 10.1006/viro.1996.0068. [DOI] [PubMed] [Google Scholar]
- 8.Vidaver A K, Koski R K, Van Etten J L. J Virol. 1973;11:799–805. doi: 10.1128/jvi.11.5.799-805.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Day L A, Mindich L. Virology. 1980;103:376–385. doi: 10.1016/0042-6822(80)90196-8. [DOI] [PubMed] [Google Scholar]
- 10.Gottlieb P, Strassman J, Qiao X, Frucht A, Mindich L. J Bacteriol. 1990;172:5774–5782. doi: 10.1128/jb.172.10.5774-5782.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottlieb P, Strassman J, Frucht A, Qiao X, Mindich L. Virology. 1991;181:589–594. doi: 10.1016/0042-6822(91)90892-f. [DOI] [PubMed] [Google Scholar]
- 12.Qiao X, Qiao J, Mindich L. J Virol. 1995;69:2926–2931. doi: 10.1128/jvi.69.5.2926-2931.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Frilander M, Bamford D H. J Mol Biol. 1995;246:418–428. doi: 10.1006/jmbi.1994.0096. [DOI] [PubMed] [Google Scholar]
- 14.Gottlieb P, Qiao X, Strassman J, Frilander M, Mindich L. Virology. 1994;200:42–47. doi: 10.1006/viro.1994.1160. [DOI] [PubMed] [Google Scholar]
- 15.Frilander M, Gottlieb P, Strassman J, Bamford D H, Mindich L. J Virol. 1992;66:5013–5017. doi: 10.1128/jvi.66.8.5013-5017.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mindich L, Qiao X, Qiao J. Virology. 1995;212:213–217. doi: 10.1006/viro.1995.1470. [DOI] [PubMed] [Google Scholar]
- 17.Onodera S, Qiao X, Qiao J, Mindich L. Virology. 1995;212:204–212. doi: 10.1006/viro.1995.1469. [DOI] [PubMed] [Google Scholar]
- 18.Mindich L, Abelson R D. Virology. 1980;103:386–391. doi: 10.1016/0042-6822(80)90197-x. [DOI] [PubMed] [Google Scholar]
- 19.Koonin E V, Gorbalenya E E, Chumakov K M. FEBS Lett. 1989;252:42–46. doi: 10.1016/0014-5793(89)80886-5. [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb P, Strassman J, Mindich L. J Virol. 1992;66:6220–6222. doi: 10.1128/jvi.66.10.6220-6222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paatero A O, Syvaoja J E, Bamford D H. J Virol. 1995;69:6729–6734. doi: 10.1128/jvi.69.11.6729-6734.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Juuti J J, Bamford D H. J Mol Biol. 1995;249:545–554. doi: 10.1006/jmbi.1995.0317. [DOI] [PubMed] [Google Scholar]
- 23.Casini G, Qiao X, Mindich L. Virology. 1994;204:251–253. doi: 10.1006/viro.1994.1529. [DOI] [PubMed] [Google Scholar]
- 24.Qiao X, Qiao J, Mindich L. J Virol. 1995;69:5502–5505. doi: 10.1128/jvi.69.9.5502-5505.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yanisch-Perron C, Vieira J, Messing J. Gene. 1985;33:103–119. doi: 10.1016/0378-1119(85)90120-9. [DOI] [PubMed] [Google Scholar]
- 26.Gottlieb P, Metzger S, Romantschuk M, Carton J, Strassman J, Bamford D H, Kalkkinen N, Mindich L. J Virol. 1988;62:181–187. doi: 10.1128/jvi.62.4.1180-1185.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Olkkonen V M, Gottlieb P, Strassman J, Qiao X, Bamford D H, Mindich L. Proc Natl Acad Sci USA. 1990;87:9173–9177. doi: 10.1073/pnas.87.23.9173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Maniatis T, Fritsch E F, Sambrook J. Molecular Cloning: A Laboratory Manual. Plainview, NY: Cold Spring Harbor Lab. Press; 1982. [Google Scholar]
- 29.Meador J, Cannon B, Cannistraro V J, Kennell D. Eur J Biochem. 1990;187:549–553. doi: 10.1111/j.1432-1033.1990.tb15336.x. [DOI] [PubMed] [Google Scholar]
- 30.Studier F W. J Mol Biol. 1973;79:237–248. doi: 10.1016/0022-2836(73)90003-x. [DOI] [PubMed] [Google Scholar]
- 31.Aebi U, Bijlenga R, Van der Broek J, Van der Broek R, Eiserling F, Kellenberger C, Kellenberger E, Mesyanzihinov V, Muller L, Showe M, Smith R, Steven A. J Supramol Struct. 1974;2:253–275. doi: 10.1002/jss.400020218. [DOI] [PubMed] [Google Scholar]
- 32.Steven A C, Bauer A C, Bisher M E, Robey F A, Black L W. J Struct Biol. 1991;106:221–236. doi: 10.1016/1047-8477(91)90072-5. [DOI] [PubMed] [Google Scholar]
- 33.Gottlieb P, Strassman J, Qiao X, Frilander M, Frucht A, Mindich L. J Virol. 1992;66:2611–2616. doi: 10.1128/jvi.66.5.2611-2616.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Frilander M, Poranen M, Bamford D H. RNA. 1995;1:510–518. [PMC free article] [PubMed] [Google Scholar]
- 35.Chen D, Zeng C Q, Wentz M J, Gorziglia M, Estes M K, Ramig R F. J Virol. 1994;68:7030–7039. doi: 10.1128/jvi.68.11.7030-7039.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Desselberger U. In: Advances in Virus Research. Maramorosch K, Murphy F A, Shatkin A J, editors. New York: Academic; 1996. pp. 69–95. [Google Scholar]
- 37.Anzola J V, Xu Z, Asamizu T, Nuss D L. Proc Natl Acad Sci USA. 1987;84:8301–8305. doi: 10.1073/pnas.84.23.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zou S, Brown E G. Virology. 1992;186:377–388. doi: 10.1016/0042-6822(92)90003-8. [DOI] [PubMed] [Google Scholar]