Abstract
The Drosophila clock genes period (per) and timeless (tim) have been studied behaviorally and biochemically, but to date there has been no viable culture system for studying the cell biology of the Drosophila clock. We have cultured pupal ring glands attached to the central nervous system and observed rhythms of period gene expression in the prothoracic gland for 4–7 days. A daily rhythm of Per protein can be entrained by light in culture, even when neural activity is blocked by tetrodotoxin. In cultures maintained for a week in constant darkness, a per-luciferase reporter gene revealed circadian rhythms of bioluminescence. As the first circadian culture system from Drosophila, the prothoracic gland provides unique advantages for investigating the interactions between clock genes and cellular physiology.
Keywords: circadian rhythms, clock gene, ring gland, prothoracic gland, luciferase reporting
Recent breakthroughs in defining the molecular components of biological clocks, most notably in Drosophila melanogaster (1–3) and Neurospora crassa (4, 5), originated with genetic screens for mutations that altered overt circadian rhythms (6–8). Two such rhythms in Drosophila are the locomotor activity rhythms of adult flies (6, 9) and the circadian gating of eclosion at the end of metamorphosis (6, 10).
The circadian system that controls the eclosion and locomotor activity rhythms of Drosophila requires the interactions of at least two genes, period (per) (6, 11, 12) and timeless (tim) (8, 13). Daily rhythms of per and tim expression appear to constitute the primary gears in the fly’s molecular circadian clock. Their transcripts accumulate during the day and decrease in abundance in the middle of the night (14, 15). Per and Tim protein levels decline during the day, and increase during the night (16–20). These molecular rhythms are thought to result from a negative feedback mechanism whereby Per and Tim proteins interact, enter the nucleus, and ultimately inhibit the transcription of their own genes (14, 18, 21–24). Light causes rapid destruction of Tim protein and may thereby set the phase of the molecular rhythms (19, 20, 25, 26).
While it is clear that these molecular interactions are required for overt behavioral rhythms, the cellular mechanisms by which clock genes mediate their behavioral effects are obscure. We now describe a tissue culture system from Drosophila that provides unique advantages for studying the effects of clock genes on cellular physiology. The pupal prothoracic gland, cultured as a complex with the central nervous system (CNS) for up to 7 days, displays rhythms of per expression that can be entrained by light. Because these rhythms persist in the presence of tetrodotoxin (TTX), they appear to be expressed endogenously by the prothoracic gland cells. Circadian rhythms of per expression were recorded in constant darkness from preparations dissected from a per-luciferase reporter strain. The results indicate that the Drosophila prothoracic glands have all the components necessary to turn on per transcription, establish the autoregulatory feedback loop, and be entrained by environmental signals, suggesting that this endocrine tissue can function, in culture, as an autonomous circadian clock.
MATERIALS AND METHODS
Animal Rearing.
Flies were raised in standard medium (27) at 25°C in 12-hr light/12-hr dark cycles (LD). White prepupae were selected at lights-on and aged in humid chambers.
Tissue Culturing.
The CNS and attached ring gland (RG) were dissected from white prepupae during the first 2 hr after lights-on. They were cultured in Schneider’s medium (GIBCO/BRL) supplemented with 20% fetal bovine serum (GIBCO/BRL), 500 ng/ml insulin, 100 units/ml penicillin, 100 μg/ml streptomycin, 20 μg/ml gentamycin, 0.25 μg/ml fungizone (28), and in some cases, 300 nM TTX (Calbiochem). They were incubated in LD at 25°C ± 1°C. Medium was replaced every 36 hr.
Anti-Per Immunostaining.
White-eyed flies (w and w per01) were used for sectioning experiments to eliminate the autofluorescence of eye pigments. Pupae were mounted in Tissue-Tek OCT compound (Miles) and frozen with finely powdered dry ice. Cryosections (8 μm) were fixed with buffered 4% paraformaldehyde and stained with affinity purified rabbit antibody to Per S-peptide (29) and Cy3-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch), diluted 1/1000. Incubations were performed for 1 hr at room temperature. Sections were counterstained with 2 μg/ml 4,6-diamidino-2-phenylindole (DAPI, Molecular Probes) in PBS and mounted in Fluoromount (Fisher) with 25 mg/ml 1,4-diazabicyclo[2.2.2]octane (DABCO; Aldrich).
Cultured CNS–RG preparations were fixed with buffered 4% formaldehyde and stained as whole mounts (30). The primary antibody was diluted in 50 mM Tris·HCl (pH 7), 0.15 M NaCl, 0.5% Triton X-100, and 2% goat serum, and incubated for 3 hr at room temperature. Cy3-conjugated goat anti-rabbit IgG (Jackson ImmunoResearch) was diluted 1/500 and incubated for 1 hr at room temperature. Whole mounts were counterstained with DAPI and mounted in Fluoromount with DABCO.
Analysis of Immunofluorescence.
Stained tissues were viewed on a Zeiss Axioskop equipped with Cy3 and DAPI filter sets (Chroma Technology, Brattleboro, VT). The intensity of Per immunofluorescence in prothoracic gland nuclei of sectioned pupae was measured in digital images acquired under standardized conditions with a Star I cooled charged-coupled device (CCD) camera (Photometrics, Tucson, AZ). Paired images of Cy3 and DAPI fluorescence were analyzed with iplab software (Signal Analytics, Vienna, VA) on a Macintosh Quadra 800. The boundaries of the nuclei of prothoracic gland cells were defined in the DAPI image of each pair, then superimposed on the Cy3 image, from which the average intensity of nuclear pixels was computed. Intensity/pixel values from 1–6 sections were averaged to obtain a value of nuclear Per immunofluorescence for each pupa.
Per immunofluorescence in the prothoracic glands of cultured CNS–RG whole mount preparations was scored blind. Prothoracic glands were identified (by DAPI staining) as a cluster of very large nuclei, then anti-Per staining in these nuclei was scored on a scale of 0–4. Glands with scores of 3–4 were defined to have “strong nuclear Per staining” for calculating the percentages in Table 1.
Table 1.
Per cycles and is entrained by light independently of neural activity in prothoracic glands of cultured CNS-RG complexes
| Percentage of cultured preparations with strong nuclear Per signal in prothoracic glands | |
Tissues were dissected from white prepupae reared at 25°C in LD. Some cultures were returned to the same incubator (Standard LD); others were placed in an incubator with an opposite phase LD cycle (Reverse LD). Some were cultured in medium containing 300 nM TTX. Preparations were fixed after 24, 36, 48, 60, 72, 84, and 96 hr in culture and stained as wholemounts with anti-Per and DAPI. Results are expressed as the percentage of each group of preparations with strong nuclear Per staining in prothoracic gland cells, with the number of preparations in each group in parenthesis.
Bioluminescence Measurements.
CNS–RG complexes dissected from white prepupae bearing the per-luc reporter gene (31) were placed in individual wells of black 96-well plates (Dynatech MicroFLUOR) containing 200 μl of culture medium supplemented with 1 mM beetle luciferin (Promega). Bioluminescence was measured in constant darkness (DD) in a TopCount Microplate Scintillation Counter (Packard). Each well was counted for 17 sec every hour for 1 week. Data were plotted as counts per second. The periods of rhythmic cultures were determined by rhythm analysis software (fft-nlls) designed by M. Straume (31).
RESULTS
Per Expression in the Pupal Prothoracic Gland.
Per expression was examined in pupae throughout metamorphosis, between white puparium formation (0 hr) and eclosion (≈96 hr). The white prepupa is a short-lived stage (15–30 min) that allows for developmental synchronization of pupae (32). Pupae were reared in LD at 25°C, so that the 24-, 48-, 72-, and 96-hr time points coincided with lights-on. In sections of 72-hr pupae, Per immunofluorescence was strongest in nuclei of the prothoracic gland cells and in a few lateral neurons between the central brain and the optic lobes (Fig. 1). This pupal expression pattern is more restricted than the adult pattern, where Per is expressed in several clusters of brain neurons as well as in photoreceptors, glial cells, corpora cardiaca, and various abdominal tissues (29, 33–35).
Figure 1.
Per is expressed in the prothoracic gland and lateral neurons of pupae. (A) Per protein immunofluorescence in a horizontal section through the head and anterior thorax of a 72-hr (lights-on) w pupa reared at 25°C in LD. Large arrows indicate prothoracic gland cells and small arrows indicate lateral neurons. The cuticle shows autofluorescence. (B) DAPI fluorescence allows prothoracic glands to be identified as clusters of very large nuclei in the RG. (Bar = 100 μm.)
The prothoracic gland synthesizes and secretes ecdysone, the steroid hormone that regulates the changes in gene expression that result in metamorphosis (36). It is part of the RG, which also includes two neuroendocrine tissues, the corpora allata and corpora cardiaca. Because it has been reported that the prothoracic gland gradually degenerates during metamorphosis (37), it was important to determine the developmental profile of Per expression in these cells. Pupae sectioned and stained every 6 hr throughout the 4 days of metamorphosis revealed oscillating levels of Per protein in the nuclei of prothoracic gland cells (Fig. 2). Beginning on the second day, prothoracic gland Per levels increased during the night and declined after lights-on, in a pattern which is consistent with the phase of Per protein cycling in adult fly heads (16–18). The specificity of the staining was confirmed by examining per01 mutants, which do not express Per protein (29, 38, 39); thus, no staining was detected in prothoracic glands of w per01 pupae aged for 30, 42, or 72 hr.
Figure 2.
Oscillations of Per protein in the nuclei of prothoracic gland cells. Pupae reared at 25°C in LD were sectioned and fixed every 6 hr. After staining with anti-Per and DAPI, the intensity of Per immunofluorescence in the nuclei of prothoracic gland cells was quantitated as described. Each point represents the mean intensity/pixel (± SEM) of Per immunofluorescence in prothoracic gland nuclei of one to three animals, based on measurements from at least two sections.
Per was detected in the lateral neurons of white prepupae, and in pupae aged for 6, 30, 54, 72, 78, and 96 hr (see Fig. 1). This is consistent with the finding that lateral neurons begin to express per during larval stages (M. Kaneko, C. Helfrich-Forster, and J. C. Hall, personal communications). Per was first detected in photoreceptors and glial cells (34) on the day of eclosion (not shown).
Rhythms of Prothoracic Gland Per in Culture.
To determine whether Per expression rhythms could occur in culture, CNS–RG complexes were dissected from white prepupae and cultured in a modified Schneider’s medium (28) at 25°C under LD conditions. Based on the prediction that the phase of Per cycling would be similar to that in adult flies (i.e., trough at lights-off and peak around lights-on; see refs. 16–18), samples were fixed at times of lights-on or lights-off after 1–4 days in culture. In whole mounts stained for Per protein and nuclei, prothoracic glands were identified as clusters of very large nuclei (Fig. 3 B, D, and F) and scored for Per staining (Fig. 3 A, C, and E). In standard LD conditions, strong Per staining was first detected in prothoracic gland nuclei after 48 hr in culture (Table 1, first row). Similarly strong nuclear Per staining was observed in many preparations fixed on subsequent days at the time of lights-on (Fig. 3 A and C; Table 1, first row, 72 and 96 hr). In contrast, the vast majority of preparations fixed at lights-off time points showed negligible Per staining in prothoracic gland nuclei (Fig. 3E; Table 1, first row, 36, 60, and 84 hr). These results indicate that daily cycles of Per protein occurred in the prothoracic glands of a significant fraction of cultured CNS–RG preparations, with high levels at lights-on and low levels at lights-off. None of the 16 preparations from w per01 pupae (fixed at 72 hr or 96 hr) showed detectable Per in prothoracic gland nuclei.
Figure 3.
Per protein cycles in the prothoracic glands of cultured CNS–RG complexes. CNS–RG complexes were dissected from white prepupae reared at 25°C in LD and cultured in the same LD regimen (see Materials and Methods). Preparations were fixed on the third day at either lights-on (A and B, C and D) or lights-off (E and F), and stained as whole mounts for Per protein (A, C, and E). Nuclei were stained with DAPI (B, D, and F). A and B show a whole CNS–RG preparation, with the prothoracic gland (arrow) prominently stained for Per. C and D show an enlargement of the prothoracic gland in A and B. E and F show the absence of Per staining in a prothoracic gland fixed at lights-off. [Bars = 100 μm (A and B) and 50 μm (C–F).]
To determine whether CNS–RG complexes were sensitive to light, some were cultured in an incubator where the phase of the LD cycle was reversed. Strong nuclear Per signals were found in significant percentages of prothoracic glands fixed at the new lights-on time points (Table 1, third row, 60 and 84 hr), while none showed strong staining at the new lights-off time points (48, 72, and 96 hr), indicating that the prothoracic gland Per rhythm was entrained to the reversed LD cycle.
Because RG have neural connections to the CNS, it was important to test the requirement for neural input in rhythms of Per expression in prothoracic glands. Conduction of action potentials in peripheral nerves of Drosophila larvae is completely blocked within 20 min of exposure to 200nM TTX (40, 41). Per cycling continued in CNS–RG preparations cultured in the chronic presence of 300nM TTX and was in the same phase as control preparations in both standard and reverse LD conditions (Table 1, second and fourth rows). These results indicate that sodium-dependent action potentials in axons from the CNS are not required for either rhythms of Per expression or its entrainment by light in the prothoracic gland.
Real-Time Measurements of per-Driven Luciferase Activity in Culture.
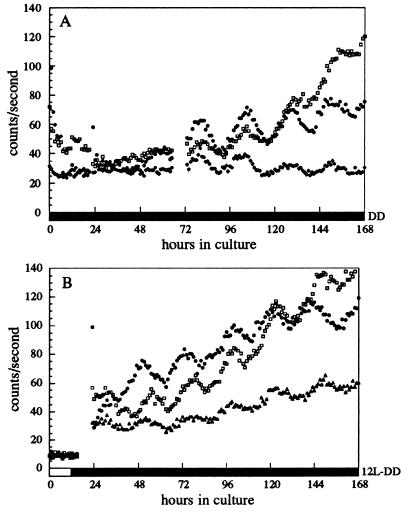
To determine whether the Per oscillations in the prothoracic gland represent a true circadian rhythm, we assayed per expression in DD using a per-luciferase reporter system. A fusion gene consisting of per regulatory sequences upstream of the firefly luciferase gene has been used to monitor per-driven bioluminescence rhythms in living flies (31). Because luciferase has a rapid rate of turnover, its activity in per-luc transgenic flies reflects per transcription in a noninvasive manner (31, 42). CNS–RG complexes were dissected from white prepupae of a per+;per-luc reporter strain and cultured in medium containing the luciferase substrate, beetle luciferin; bioluminescence was monitored for 1 week in DD (see Materials and Methods). Of the 60 per+;per-luc preparations monitored, 7 displayed strong per-driven luciferase activity rhythms that persisted for 4–5 days with an average period of 24.0 ± 0.3 h (Fig. 4). Thirteen others displayed either 2 days of strong oscillations or 3–6 days of weaker oscillations (data not shown). Taken together, these rhythmic preparations represent 33% of the total number monitored. Seven per01;per-luc CNS–RG complexes were monitored; none of these showed rhythms of luciferase activity even though they maintained an average luminescence level of ≈50 counts/sec (data not shown).
Figure 4.
Circadian rhythms of per-driven luciferase activity in DD in CNS–RG preparations from per-luc pupae. CNS–RG complexes from per+;per-luc white prepupae were cultured in medium containing 1 mM beetle luciferin. Emanating luminescence (counts/sec) was measured with a Packard TopCount Microplate Scintillation Counter for 1 week in DD. (A) One group of preparations was placed in DD immediately after dissection. (B) A second group was exposed to 12 hr of light before being placed in DD. Technical difficulties interrupted data collection between 65 and 73 hr in A. Blank wells had basal luminescence levels of 10 counts/sec.
The per-driven bioluminescence rhythms in different preparations were not precisely in phase with one another (Fig. 4B), confirming that they were driven by endogenous clocks as opposed to external factors. Because the prothoracic gland cells are the most prominent and largest cells that express Per in the CNS–RG complexes (see Fig. 3A), it is reasonable to conclude that the circadian rhythms of per-driven bioluminescence emanated from these cells.
DISCUSSION
Per Rhythms in Prothoracic Glands in Vivo and in Culture.
The prothoracic gland is the major site of per expression in Drosophila pupae. A daily rhythm of Per protein in the nuclei of prothoracic gland cells begins during the second day of metamorphosis and continues until the cells degrade around the time of eclosion. Prothoracic gland Per levels are low at lights-off, and increase during the night, similar to the phase of Per protein cycling in adults (16–18). Per protein was also observed in lateral neurons of pupae, but not in photoreceptors or glial cells, which abundantly express per in adult flies.
The prothoracic glands of CNS–RG complexes sustained rhythms of per expression in culture for many days in both LD and DD conditions. The rhythm was entrained to oppositely phased LD cycles, demonstrating that the preparations are sensitive to light. These results were unaffected by TTX, indicating that sodium action potentials in nerves connecting the RG to the CNS are not required for either the rhythm of prothoracic gland Per or its entrainment by light. Axons in these nerves originate in brain neurosecretory cells. Although the cell bodies of insect neurosecretory cells can sustain calcium action potentials, their spontaneous activity and axonal spikes are blocked by TTX (43, 44). Thus, it is unlikely that the RG receives electrical signals from the CNS in the presence of TTX. Circadian oscillations in rat suprachiasmatic nuclei also continue in the presence of TTX, although entrainment signals from the retina are blocked (45–47). Our results suggest that Drosophila prothoracic glands can mediate both entrainment and pacemaker functions in the absence of neural input.
Prothoracic Gland Per and Eclosion Rhythms.
The prothoracic glands function to produce the steroid hormone ecdysone. Circadian rhythms in ecdysteroid levels have been measured in the hemolymph of the blood sucking insect Rhodnius prolixus (48) and the wax moth Galleria mellonella (49). Rhodnius prothoracic glands display circadian rhythms of ecdysteroid synthesis (50) and are sensitive to light (51), as are prothoracic glands of the moth Samia cynthia ricini (52).
In Drosophila pupae, ecdysteroid titers peak on the second day of metamorphosis (36), although there is no direct evidence for their circadian modulation. The present evidence for rhythmic clock gene expression in prothoracic glands throughout metamorphosis suggests that ecdysteroid release might vary circadianly in Drosophila pupae. Such a rhythmic release of ecdysteroids could contribute to the circadian gating of eclosion. Truman proposed many years ago (53) that the timing of eclosion in silkmoths is determined by the interactions between two circadian clocks: one located in the brain that produces a daily gate for release of eclosion hormone, and another outside the brain that produces a daily modulation of ecdysteroid levels (53–55). This suggestion echoed Pittendrigh’s (56) conclusion, based on extensive studies of eclosion rhythms in Drosophila pseudoobscura, that these rhythms are produced by the interactions between a central pacemaker and a slave oscillator.
Thus, we must consider the hypothesis that rhythmic expression of per in pupal prothoracic glands is a cellular manifestation of one of a pair of oscillators that interact to control the circadian gating of eclosion. Preliminary data involving a particular transgenic strain suggest that such a circadian function for prothoracic glands is plausible. Adult flies of the 7.2:2 transgenic strain express per only in lateral brain neurons; expression in these cells is sufficient to generate locomotor activity rhythms (35). In prothoracic glands of 7.2:2 pupae, however, Per signal is absent (I.F.E., unpublished results), and this strain does not display circadian rhythms of eclosion (M. J. Hamblen and J. C. Hall, personal communication; Y. Cheng and P. E. Hardin, personal communication). Although preliminary, these results support a role for prothoracic gland Per in the generation of eclosion rhythms.
Prothoracic Glands as a Circadian Culture System.
Results from cultured CNS–RG complexes in both LD (Table 1) and DD conditions (Fig. 4) suggest that the clock is running in some variable fraction of the preparations. This variability would be problematic if further studies required the synchrony of many cultures; with luciferase as a real-time reporter of per expression, however, clock function can be monitored in individual preparations. Thus, one can easily identify individual cultures that are rhythmic, and estimate the rhythm’s phase at the time of any experimental manipulation. By combining the per-luc reporter gene with mutations disrupting specific signal transduction molecules, for example, the pathways by which light and temperature cues are transduced into effects on clock gene expression can be defined. Similar genetic strategies can be used to define pathways by which clock genes influence cellular membrane properties or ecdysone release from cultured prothoracic glands.
Circadian rhythms of neural activity or hormone release have been studied in other circadian culture systems. For example, chicken pineal cells (57–60), Xenopus retinal photoreceptors (61), and mammalian retinas (62) display circadian rhythms of melatonin release in culture. Neurons in the molluscan basal retina (63–65) and in rat suprachiasmatic nuclei (47, 66, 67) express circadian firing rhythms. Studies of these culture systems have illuminated a diversity of mechanisms by which light and neurotransmitters (i.e., inputs to the clock) can influence circadian physiology (i.e., clock output).
Similar studies in Drosophila prothoracic glands will allow simultaneous analysis of physiological activity (e.g., membrane properties or ecdysone release) and pacemaker activity in the form of clock gene expression. To our knowledge, this is the first circadian culture system from an animal with extensive clock genetics, and as such, it offers unique advantages for genetic analyses of clock cell biology. Its ability to express an entrainable circadian rhythm of per expression indicates that it contains all the components of a working biological clock, including molecules involved in maintaining the per gene regulatory feedback loop and its photoentrainment. Use of the per-luciferase reporter system will allow direct monitoring of the state of the clock in individual cultures. Pacemaker activity, entrainment transduction pathways, and cellular physiology can be probed in the same cells, providing a system that is uniquely amenable to defining the cellular mechanisms by which clock genes regulate behavioral rhythmicity.
Acknowledgments
We thank J. C. Hall and S. Gilbert for discussion and critical comments on the manuscript, and M. Hamblen, J. Hall, M. Kaneko, C. Helfrich-Forster, Y. Cheng, and P. E. Hardin for sharing unpublished results. This work was supported by a National Science Foundation Postdoctoral Fellowship (I.F.E.), National Science Foundation Grants IBN-9310256 and IBN-9057703 (K.K.S.), and National Institutes of Health Grants GM-33205 and MH-51573 to J. C. Hall.
ABBREVIATIONS
- CNS
central nervous system
- RG
ring gland
- LD
12-hr light/12-hr dark cycles
- DD
constant darkness
- TTX
tetrodotoxin
- DAPI
4,6-diamidino-2-phenylindole
References
- 1.Hall J C. Trends Neurosci. 1995;18:230–240. doi: 10.1016/0166-2236(95)93908-g. [DOI] [PubMed] [Google Scholar]
- 2.Hardin P, Siwicki K K. Semin Neurosci. 1995;7:15–25. [Google Scholar]
- 3.Sehgal A. Semin Neurosci. 1995;7:27–35. [Google Scholar]
- 4.Aronson B D, Johnson K A, Dunlap J C. Proc Natl Acad Sci USA. 1994;91:7683–7687. doi: 10.1073/pnas.91.16.7683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Loros J. Semin Neurosci. 1995;7:3–13. [Google Scholar]
- 6.Konopka R J, Benzer S. Proc Natl Acad Sci USA. 1971;68:2112–2146. doi: 10.1073/pnas.68.9.2112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Feldman J F, Hoyle M. Genetics. 1973;75:605–613. doi: 10.1093/genetics/75.4.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sehgal A, Price J L, Man B, Young M W. Science. 1994;263:1603–1606. doi: 10.1126/science.8128246. [DOI] [PubMed] [Google Scholar]
- 9.Hamblen-Coyle M J, Wheeler D A, Rutila J E, Rosbash M, Hall J C. J Insect Behav. 1992;5:417–446. [Google Scholar]
- 10.Pittendrigh C S, Skopik S D. Proc Natl Acad Sci USA. 1970;65:500–507. doi: 10.1073/pnas.65.3.500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson F R, Bargiello T A, Yun S-H, Young M W. Nature (London) 1986;320:185–188. doi: 10.1038/320185a0. [DOI] [PubMed] [Google Scholar]
- 12.Citri Y, Colot H V, Jacquier A C, Yu Q, Hall J C, Baltimore D, Rosbash M. Nature (London) 1987;326:42–47. doi: 10.1038/326042a0. [DOI] [PubMed] [Google Scholar]
- 13.Myers M P, Wager-Smith K, Wesley C S, Young M W, Sehgal A. Science. 1995;270:805–808. doi: 10.1126/science.270.5237.805. [DOI] [PubMed] [Google Scholar]
- 14.Hardin P E, Hall J C, Rosbash M. Nature (London) 1990;343:536–540. doi: 10.1038/343536a0. [DOI] [PubMed] [Google Scholar]
- 15.Sehgal A, Rothenfluh-Hilfiker A, Hunter-Ensor M, Chen Y, Myers M P, Young M W. Science. 1995;270:808–810. doi: 10.1126/science.270.5237.808. [DOI] [PubMed] [Google Scholar]
- 16.Zerr D M, Rosbash M, Hall J C, Siwicki K K. J Neurosci. 1990;10:2749–2762. doi: 10.1523/JNEUROSCI.10-08-02749.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Edery I, Zwiebel L J, Dembinska M E, Rosbash M. Proc Natl Acad Sci USA. 1994;91:2260–2264. doi: 10.1073/pnas.91.6.2260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng H, Hardin P E, Rosbash M. EMBO J. 1994;13:3590–3598. doi: 10.1002/j.1460-2075.1994.tb06666.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hunter-Ensor M, Ousley A, Sehgal A. Cell. 1996;84:677–685. doi: 10.1016/s0092-8674(00)81046-6. [DOI] [PubMed] [Google Scholar]
- 20.Myers M P, Wager-Smith K, Rothenfluh-Hilfiker A, Young M W. Science. 1996;271:1736–1740. doi: 10.1126/science.271.5256.1736. [DOI] [PubMed] [Google Scholar]
- 21.Hardin P E, Hall J C, Rosbash M. Proc Natl Acad Sci USA. 1992;89:11711–11715. doi: 10.1073/pnas.89.24.11711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtin K D, Huang Z J, Rosbash M. Neuron. 1995;14:365–372. doi: 10.1016/0896-6273(95)90292-9. [DOI] [PubMed] [Google Scholar]
- 23.Gekakis N, Saez L, Delahaye-Brown A-M, Myers M P, Sehgal A, Young M W, Weitz C J. Science. 1995;270:811–815. doi: 10.1126/science.270.5237.811. [DOI] [PubMed] [Google Scholar]
- 24.Saez L, Young M W. Neuron. 1996;17:911–920. doi: 10.1016/s0896-6273(00)80222-6. [DOI] [PubMed] [Google Scholar]
- 25.Zeng H, Qian Z, Myers M P, Rosbash M. Nature (London) 1996;380:129–135. doi: 10.1038/380129a0. [DOI] [PubMed] [Google Scholar]
- 26.Lee C, Parikh V, Itsukaichi T, Bae K, Edery I. Science. 1996;271:1740–1744. doi: 10.1126/science.271.5256.1740. [DOI] [PubMed] [Google Scholar]
- 27.Elgin S R, Miller D W. In: The Genetics and Biology of Drosophila. Ashburner M, Wright T R F, editors. 2A. New York: Academic; 1978. pp. 112–121. [Google Scholar]
- 28.Saito M, Wu C-F. J Neurosci. 1991;11:2135–2150. doi: 10.1523/JNEUROSCI.11-07-02135.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Siwicki K K, Eastman C, Petersen G, Rosbash M, Hall J C. Neuron. 1988;1:141–150. doi: 10.1016/0896-6273(88)90198-5. [DOI] [PubMed] [Google Scholar]
- 30.Boyd L, O’Toole E, Thummel C S. Development (Cambridge, UK) 1991;112:981–995. doi: 10.1242/dev.112.4.981. [DOI] [PubMed] [Google Scholar]
- 31.Brandes C, Plautz J D, Stanewsky R, Jamison C F, Straume M, Wood K V, Kay S A, Hall J C. Neuron. 1996;16:687–692. doi: 10.1016/s0896-6273(00)80088-4. [DOI] [PubMed] [Google Scholar]
- 32.Bainbridge S P, Bownes M. J Embryol Exp Morphol. 1981;66:57–80. [PubMed] [Google Scholar]
- 33.Liu X, Lorenz L, Yu Q, Hall J C, Rosbash M. Genes Dev. 1988;2:228–238. doi: 10.1101/gad.2.2.228. [DOI] [PubMed] [Google Scholar]
- 34.Ewer J, Frisch B, Hamblen-Coyle M J, Rosbash M, Hall J C. J Neurosci. 1992;12:3321–3349. doi: 10.1523/JNEUROSCI.12-09-03321.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Frisch B, Hardin P E, Hamblen-Coyle M J, Rosbash M, Hall J C. Neuron. 1994;12:555–570. doi: 10.1016/0896-6273(94)90212-7. [DOI] [PubMed] [Google Scholar]
- 36.Riddiford L M. In: The Development of Drosophila Melanogaster. Bate M, Martinez Arias A, editors. Vol. 2. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 899–939. [Google Scholar]
- 37.Dai J, Henrich V, Gilbert L. Cell Tissue Res. 1991;265:435–445. doi: 10.1007/BF00340866. [DOI] [PubMed] [Google Scholar]
- 38.Yu Q, Jacquier A C, Citri Y, Hamblen M, Hall J C, Rosbash M. Proc Natl Acad Sci USA. 1987;84:784–788. doi: 10.1073/pnas.84.3.784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baylies M K, Bargiello T A, Jackson F R, Young M W. Nature (London) 1987;326:390–392. doi: 10.1038/326390a0. [DOI] [PubMed] [Google Scholar]
- 40.Gitschier J, Strichartz G R, Hall L M. Biochim Biophys Acta. 1980;595:291–303. doi: 10.1016/0005-2736(80)90091-7. [DOI] [PubMed] [Google Scholar]
- 41.Wu C-F, Ganetsky B. Nature (London) 1980;286:814–816. doi: 10.1038/286814a0. [DOI] [PubMed] [Google Scholar]
- 42.Wood K V. Curr Opin Biotechnol. 1995;6:50–58. doi: 10.1016/0958-1669(95)80009-3. [DOI] [PubMed] [Google Scholar]
- 43.Carrow G M, Calabrese R L, Williams C M. J Neurosci. 1984;4:1034–1044. doi: 10.1523/JNEUROSCI.04-04-01034.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miyazaki S. J Comp Physiol A. 1980;140:43–52. [Google Scholar]
- 45.Schwartz W J, Gross R A, Morton M T. Proc Natl Acad Sci USA. 1987;84:1694–1698. doi: 10.1073/pnas.84.6.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schwartz W J. J Biol Rhythms. 1991;6:149–158. doi: 10.1177/074873049100600205. [DOI] [PubMed] [Google Scholar]
- 47.Welsh D K, Logothetis D E, Meister M, Reppert S M. Neuron. 1995;14:697–706. doi: 10.1016/0896-6273(95)90214-7. [DOI] [PubMed] [Google Scholar]
- 48.Ampleford E J, Steel C G H. Gen Comp Endocrinol. 1985;59:453–459. doi: 10.1016/0016-6480(85)90404-6. [DOI] [PubMed] [Google Scholar]
- 49.Cymborowski B, Smietanko A, Delbecque J P. Comp Biochem Physiol A. 1989;94:431–438. [Google Scholar]
- 50.Vafopoulou X, Steel C G H. Gen Comp Endocrinol. 1991;83:27–34. doi: 10.1016/0016-6480(91)90102-c. [DOI] [PubMed] [Google Scholar]
- 51.Vafopoulou X, Steel C G H. Gen Comp Endocrinol. 1992;86:1–9. doi: 10.1016/0016-6480(92)90119-5. [DOI] [PubMed] [Google Scholar]
- 52.Mizoguchi A, Ishizaki H. Proc Natl Acad Sci USA. 1982;79:2726–2730. doi: 10.1073/pnas.79.8.2726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Truman J W. In: Photoperiodic Regulation of Insect and Molluscan Hormones. Porter R, Collins G M, editors. London: Pitman; 1984. pp. 221–239. [Google Scholar]
- 54.Truman J W, Rountree D B, Reiss S E, Schwartz L M. J Insect Physiol. 1983;29:895–900. [Google Scholar]
- 55.Schwartz L M, Truman J W. Dev Biol. 1983;99:103–114. doi: 10.1016/0012-1606(83)90257-9. [DOI] [PubMed] [Google Scholar]
- 56.Pittendrigh C S. In: Biological Clocks in Seasonal Reproductive Cycles. Follett B K, Follett D E, editors. New York: Wiley; 1981. pp. 1–35. [Google Scholar]
- 57.Deguchi T A. Nature (London) 1979;282:94–96. doi: 10.1038/282094a0. [DOI] [PubMed] [Google Scholar]
- 58.Zatz M, Mullen D A, Moskal J R. Brain Res. 1988;438:199–215. doi: 10.1016/0006-8993(88)91339-x. [DOI] [PubMed] [Google Scholar]
- 59.Robertson L M, Takahashi J S. J Neurosci. 1988;8:12–21. doi: 10.1523/JNEUROSCI.08-01-00012.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pickard G E, Tang W X. Brain Res. 1993;627:141–146. doi: 10.1016/0006-8993(93)90757-e. [DOI] [PubMed] [Google Scholar]
- 61.Cahill G M, Besharse J C. Neuron. 1993;10:573–577. doi: 10.1016/0896-6273(93)90160-s. [DOI] [PubMed] [Google Scholar]
- 62.Tosini G, Menaker M. Science. 1996;272:419–421. doi: 10.1126/science.272.5260.419. [DOI] [PubMed] [Google Scholar]
- 63.Jacklet J W. Science. 1969;164:257–262. doi: 10.1126/science.164.3879.562. [DOI] [PubMed] [Google Scholar]
- 64.Block G D, Wallace S. Science. 1982;217:155–157. doi: 10.1126/science.217.4555.155. [DOI] [PubMed] [Google Scholar]
- 65.Michel S, Guesz M E, Zaritsky J J, Block G D. Science. 1993;259:239–241. doi: 10.1126/science.8421785. [DOI] [PubMed] [Google Scholar]
- 66.Green D J, Gillette R. Brain Res. 1982;245:198–200. doi: 10.1016/0006-8993(82)90361-4. [DOI] [PubMed] [Google Scholar]
- 67.Gillette M U. In: Suprachiasmatic Nucleus: The Mind’s Clock. Klein D C, Moore R Y, Reppert S M, editors. New York: Oxford Univ. Press; 1991. pp. 125–143. [Google Scholar]