Abstract
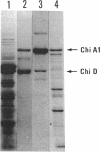
The gene (chiD) encoding the precursor of chitinase D was found to be located immediately upstream of the chiA gene, encoding chitinase A1, which is a key enzyme in the chitinase system of Bacillus circulans WL-12. Sequencing analysis revealed that the deduced polypeptide encoded by the chiD gene was 488 amino acids long and the distance between the coding regions of the chiA and chiD genes was 103 bp. Remarkable similarity was observed between the N-terminal one-third of chitinase D and the C-terminal one-third of chitinase A1. The N-terminal 47-amino-acid segment (named ND) of chitinase D showed a 61.7% amino acid match with the C-terminal segment (CA) of chitinase A1. The following 95-amino-acid segment (R-D) of chitinase D showed 62.8 and 60.6% amino acid matches, respectively, to the previously reported type III-like repeating units R-1 and R-2 in chitinase A1, which were shown to be homologous to the fibronectin type III sequence. A 73-amino-acid segment (residues 247 to 319) located in the putative activity domain of chitinase D was found to show considerable sequence similarity not only to other bacterial chitinases and class III higher-plant chitinases but also to Streptomyces plicatus endo-beta-N-acetylglucosaminidase H and the Kluyveromyces lactis killer toxin alpha subunit. The evolutionary and functional meanings of these similarities are discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- ARTMAN M. The production of hydrogen sulphide from thiosulphate by Escherichia coli. J Gen Microbiol. 1956 Apr;14(2):315–322. doi: 10.1099/00221287-14-2-315. [DOI] [PubMed] [Google Scholar]
- Ames G. F. Resolution of bacterial proteins by polyacrylamide gel electrophoresis on slabs. Membrane, soluble, and periplasmic fractions. J Biol Chem. 1974 Jan 25;249(2):634–644. [PubMed] [Google Scholar]
- Harpster M. H., Dunsmuir P. Nucleotide sequence of the chitinase B gene of Serratia marcescens QMB1466. Nucleic Acids Res. 1989 Jul 11;17(13):5395–5395. doi: 10.1093/nar/17.13.5395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirano S., Ohe Y., Ono H. Selective N-acylation of chitosan. Carbohydr Res. 1976 Apr;47(2):315–320. doi: 10.1016/s0008-6215(00)84198-1. [DOI] [PubMed] [Google Scholar]
- Jones F. S., Hoffman S., Cunningham B. A., Edelman G. M. A detailed structural model of cytotactin: protein homologies, alternative RNA splicing, and binding regions. Proc Natl Acad Sci U S A. 1989 Mar;86(6):1905–1909. doi: 10.1073/pnas.86.6.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones J. D., Grady K. L., Suslow T. V., Bedbrook J. R. Isolation and characterization of genes encoding two chitinase enzymes from Serratia marcescens. EMBO J. 1986 Mar;5(3):467–473. doi: 10.1002/j.1460-2075.1986.tb04235.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamei K., Yamamura Y., Hara S., Ikenaka T. Amino acid sequence of chitinase from Streptomyces erythraeus. J Biochem. 1989 Jun;105(6):979–985. doi: 10.1093/oxfordjournals.jbchem.a122791. [DOI] [PubMed] [Google Scholar]
- Kornblihtt A. R., Umezawa K., Vibe-Pedersen K., Baralle F. E. Primary structure of human fibronectin: differential splicing may generate at least 10 polypeptides from a single gene. EMBO J. 1985 Jul;4(7):1755–1759. doi: 10.1002/j.1460-2075.1985.tb03847.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LUNT M. R., KENT P. W. A chitinase system from Carcinus maenas. Biochim Biophys Acta. 1960 Nov 4;44:371–373. doi: 10.1016/0006-3002(60)91581-x. [DOI] [PubMed] [Google Scholar]
- Laemmli U. K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970 Aug 15;227(5259):680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Malcolm B. A., Rosenberg S., Corey M. J., Allen J. S., de Baetselier A., Kirsch J. F. Site-directed mutagenesis of the catalytic residues Asp-52 and Glu-35 of chicken egg white lysozyme. Proc Natl Acad Sci U S A. 1989 Jan;86(1):133–137. doi: 10.1073/pnas.86.1.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. A genetic approach to analyzing membrane protein topology. Science. 1986 Sep 26;233(4771):1403–1408. doi: 10.1126/science.3529391. [DOI] [PubMed] [Google Scholar]
- Metraux J. P., Burkhart W., Moyer M., Dincher S., Middlesteadt W., Williams S., Payne G., Carnes M., Ryals J. Isolation of a complementary DNA encoding a chitinase with structural homology to a bifunctional lysozyme/chitinase. Proc Natl Acad Sci U S A. 1989 Feb;86(3):896–900. doi: 10.1073/pnas.86.3.896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moos M., Tacke R., Scherer H., Teplow D., Früh K., Schachner M. Neural adhesion molecule L1 as a member of the immunoglobulin superfamily with binding domains similar to fibronectin. Nature. 1988 Aug 25;334(6184):701–703. doi: 10.1038/334701a0. [DOI] [PubMed] [Google Scholar]
- POWNING R. F., IRZYKIEWICZ H. STUDIES ON THE CHITINASE SYSTEM IN BEAN AND OTHER SEEDS. Comp Biochem Physiol. 1965 Jan;14:127–133. doi: 10.1016/0010-406x(65)90013-7. [DOI] [PubMed] [Google Scholar]
- Robbins P. W., Trimble R. B., Wirth D. F., Hering C., Maley F., Maley G. F., Das R., Gibson B. W., Royal N., Biemann K. Primary structure of the Streptomyces enzyme endo-beta-N-acetylglucosaminidase H. J Biol Chem. 1984 Jun 25;259(12):7577–7583. [PubMed] [Google Scholar]
- Roberts R. L., Cabib E. Serratia marcescens chitinase: one-step purification and use for the determination of chitin. Anal Biochem. 1982 Dec;127(2):402–412. doi: 10.1016/0003-2697(82)90194-4. [DOI] [PubMed] [Google Scholar]
- Samac D. A., Hironaka C. M., Yallaly P. E., Shah D. M. Isolation and Characterization of the Genes Encoding Basic and Acidic Chitinase in Arabidopsis thaliana. Plant Physiol. 1990 Jul;93(3):907–914. doi: 10.1104/pp.93.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanger F., Nicklen S., Coulson A. R. DNA sequencing with chain-terminating inhibitors. Proc Natl Acad Sci U S A. 1977 Dec;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinshi H., Neuhas J. M., Ryals J., Meins F., Jr Structure of a tobacco endochitinase gene: evidence that different chitinase genes can arise by transposition of sequences encoding a cysteine-rich domain. Plant Mol Biol. 1990 Mar;14(3):357–368. doi: 10.1007/BF00028772. [DOI] [PubMed] [Google Scholar]
- Watanabe T., Oyanagi W., Suzuki K., Tanaka H. Chitinase system of Bacillus circulans WL-12 and importance of chitinase A1 in chitin degradation. J Bacteriol. 1990 Jul;172(7):4017–4022. doi: 10.1128/jb.172.7.4017-4022.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe T., Suzuki K., Oyanagi W., Ohnishi K., Tanaka H. Gene cloning of chitinase A1 from Bacillus circulans WL-12 revealed its evolutionary relationship to Serratia chitinase and to the type III homology units of fibronectin. J Biol Chem. 1990 Sep 15;265(26):15659–15665. [PubMed] [Google Scholar]
- Yahata N., Watanabe T., Nakamura Y., Yamamoto Y., Kamimiya S., Tanaka H. Structure of the gene encoding beta-1,3-glucanase A1 of Bacillus circulans WL-12. Gene. 1990 Jan 31;86(1):113–117. doi: 10.1016/0378-1119(90)90122-8. [DOI] [PubMed] [Google Scholar]
- Yamada H., Imoto T. A convenient synthesis of glycolchitin, a substrate of lysozyme. Carbohydr Res. 1981 May 18;92(1):160–162. doi: 10.1016/s0008-6215(00)85993-5. [DOI] [PubMed] [Google Scholar]