Abstract
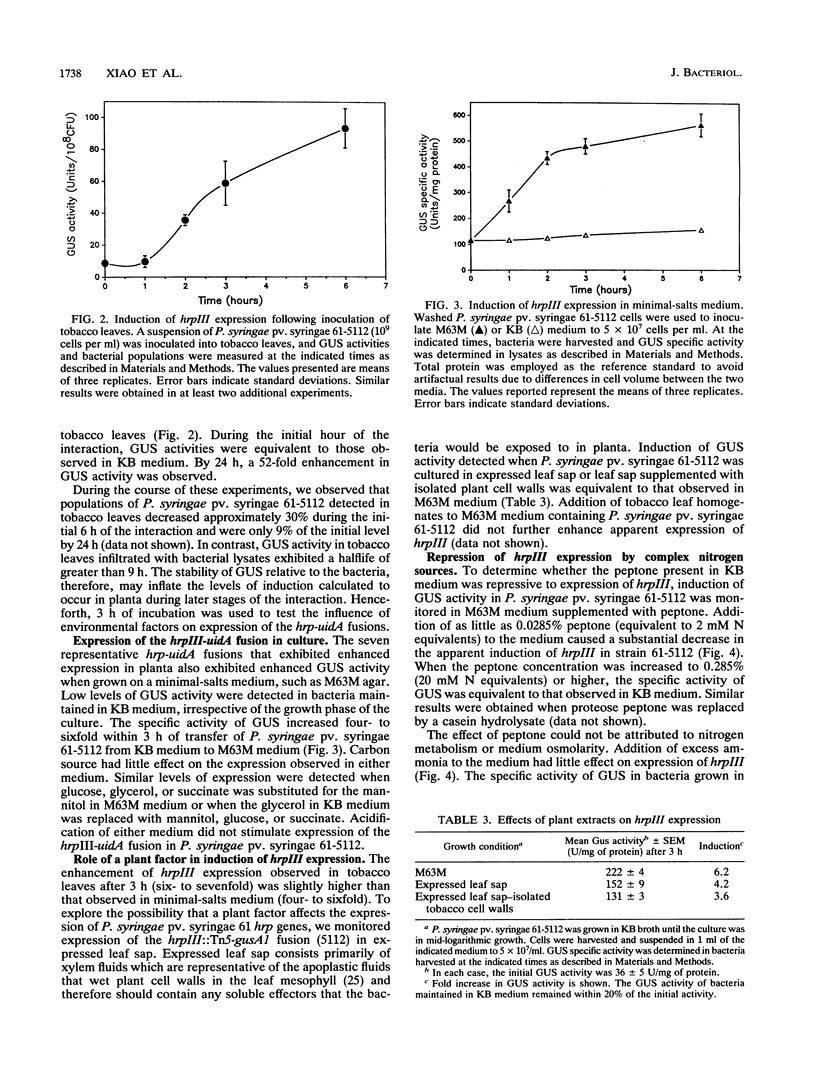
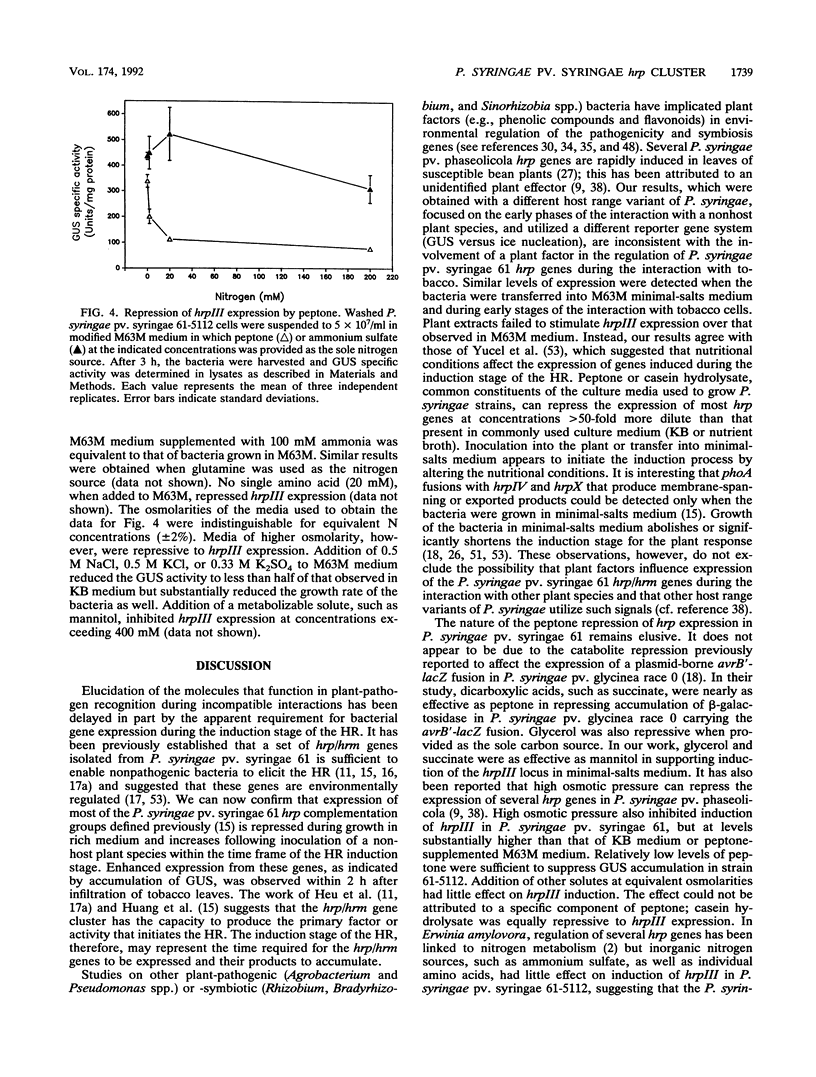
The ability of Pseudomonas syringae pv. syringae 61 to elicit the hypersensitive response in nonhost plant species has been linked to a cluster of hrp/hrm genes whose expression appears to be environmentally regulated. To understand the genetic organization of this hrp/hrm gene cluster and its expression during the interaction with nonhost plant species better, we constructed a set of chromosomal hrp-uidA fusions in P. syringae pv. syringae 61 by Tn5-gusA1 mutagenesis of the cloned hrp/hrm gene cluster and transferred them into the genome by marker exchange mutagenesis. Complementation analysis employing plasmid-borne Tn5-gusA1 insertions and previously characterized chromosomal TnphoA mutations defined at least eight apparent transcriptional units within the hrp/hrm cluster, several of which were multicistronic. The expression of hrp-uidA fusions in seven of these apparent hrp transcriptional units increased following inoculation into tobacco leaves. Enhanced expression from a representative fusion was detected 1 h after inoculation of tobacco leaves. The induction observed in planta was similar to the levels detected following culture of the bacteria in minimal-salts medium: irrespective of the carbon source. Complex amino acid sources, such as peptone, repressed the expression of P. syringae pv. syringae 61 hrp genes at levels exceeding 0.028%. The results indicate that enhanced expression of hrp genes occurs early in the interaction with nonhost plant species in an apparent response to altered nutritional conditions.
Full text
PDF





Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bradford M. M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976 May 7;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Ditta G., Stanfield S., Corbin D., Helinski D. R. Broad host range DNA cloning system for gram-negative bacteria: construction of a gene bank of Rhizobium meliloti. Proc Natl Acad Sci U S A. 1980 Dec;77(12):7347–7351. doi: 10.1073/pnas.77.12.7347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dower W. J., Miller J. F., Ragsdale C. W. High efficiency transformation of E. coli by high voltage electroporation. Nucleic Acids Res. 1988 Jul 11;16(13):6127–6145. doi: 10.1093/nar/16.13.6127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983 Jun 5;166(4):557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- Huang H. C., Schuurink R., Denny T. P., Atkinson M. M., Baker C. J., Yucel I., Hutcheson S. W., Collmer A. Molecular cloning of a Pseudomonas syringae pv. syringae gene cluster that enables Pseudomonas fluorescens to elicit the hypersensitive response in tobacco plants. J Bacteriol. 1988 Oct;170(10):4748–4756. doi: 10.1128/jb.170.10.4748-4756.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh T. V., Dahlbeck D., Staskawicz B. J. Bacterial blight of soybean: regulation of a pathogen gene determining host cultivar specificity. Science. 1989 Sep 22;245(4924):1374–1377. doi: 10.1126/science.2781284. [DOI] [PubMed] [Google Scholar]
- KING E. O., WARD M. K., RANEY D. E. Two simple media for the demonstration of pyocyanin and fluorescin. J Lab Clin Med. 1954 Aug;44(2):301–307. [PubMed] [Google Scholar]
- Keen N. T. Gene-for-gene complementarity in plant-pathogen interactions. Annu Rev Genet. 1990;24:447–463. doi: 10.1146/annurev.ge.24.120190.002311. [DOI] [PubMed] [Google Scholar]
- Li T. H., Benson S. A., Hutcheson S. W. Phenotypic expression of the Pseudomonas syringae pv. syringae 61 hrp/hrm gene cluster in Escherichia coli MC4100 requires a functional porin. J Bacteriol. 1992 Mar;174(6):1742–1749. doi: 10.1128/jb.174.6.1742-1749.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindgren P. B., Peet R. C., Panopoulos N. J. Gene cluster of Pseudomonas syringae pv. "phaseolicola" controls pathogenicity of bean plants and hypersensitivity of nonhost plants. J Bacteriol. 1986 Nov;168(2):512–522. doi: 10.1128/jb.168.2.512-522.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Long S. R. Rhizobium genetics. Annu Rev Genet. 1989;23:483–506. doi: 10.1146/annurev.ge.23.120189.002411. [DOI] [PubMed] [Google Scholar]
- Maas R. An improved colony hybridization method with significantly increased sensitivity for detection of single genes. Plasmid. 1983 Nov;10(3):296–298. doi: 10.1016/0147-619x(83)90045-8. [DOI] [PubMed] [Google Scholar]
- Manoil C., Beckwith J. TnphoA: a transposon probe for protein export signals. Proc Natl Acad Sci U S A. 1985 Dec;82(23):8129–8133. doi: 10.1073/pnas.82.23.8129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo Y. Y., Gross D. C. Plant signal molecules activate the syrB gene, which is required for syringomycin production by Pseudomonas syringae pv. syringae. J Bacteriol. 1991 Sep;173(18):5784–5792. doi: 10.1128/jb.173.18.5784-5792.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niepold F., Anderson D., Mills D. Cloning determinants of pathogenesis from Pseudomonas syringae pathovar syringae. Proc Natl Acad Sci U S A. 1985 Jan;82(2):406–410. doi: 10.1073/pnas.82.2.406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahme L. G., Mindrinos M. N., Panopoulos N. J. Genetic and transcriptional organization of the hrp cluster of Pseudomonas syringae pv. phaseolicola. J Bacteriol. 1991 Jan;173(2):575–586. doi: 10.1128/jb.173.2.575-586.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruvkun G. B., Ausubel F. M. A general method for site-directed mutagenesis in prokaryotes. Nature. 1981 Jan 1;289(5793):85–88. doi: 10.1038/289085a0. [DOI] [PubMed] [Google Scholar]
- Sharma S. B., Signer E. R. Temporal and spatial regulation of the symbiotic genes of Rhizobium meliloti in planta revealed by transposon Tn5-gusA. Genes Dev. 1990 Mar;4(3):344–356. doi: 10.1101/gad.4.3.344. [DOI] [PubMed] [Google Scholar]
- Staskawicz B., Dahlbeck D., Keen N., Napoli C. Molecular characterization of cloned avirulence genes from race 0 and race 1 of Pseudomonas syringae pv. glycinea. J Bacteriol. 1987 Dec;169(12):5789–5794. doi: 10.1128/jb.169.12.5789-5794.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winans S. C. Transcriptional induction of an Agrobacterium regulatory gene at tandem promoters by plant-released phenolic compounds, phosphate starvation, and acidic growth media. J Bacteriol. 1990 May;172(5):2433–2438. doi: 10.1128/jb.172.5.2433-2438.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yucel I., Xiao Y. X., Hutcheson S. W. Influence of Pseudomonas syringae culture conditions on initiation of the hypersensitive response of culture tobacco cells. Appl Environ Microbiol. 1989 Jul;55(7):1724–1729. doi: 10.1128/aem.55.7.1724-1729.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Bruijn F. J., Lupski J. R. The use of transposon Tn5 mutagenesis in the rapid generation of correlated physical and genetic maps of DNA segments cloned into multicopy plasmids--a review. Gene. 1984 Feb;27(2):131–149. doi: 10.1016/0378-1119(84)90135-5. [DOI] [PubMed] [Google Scholar]



